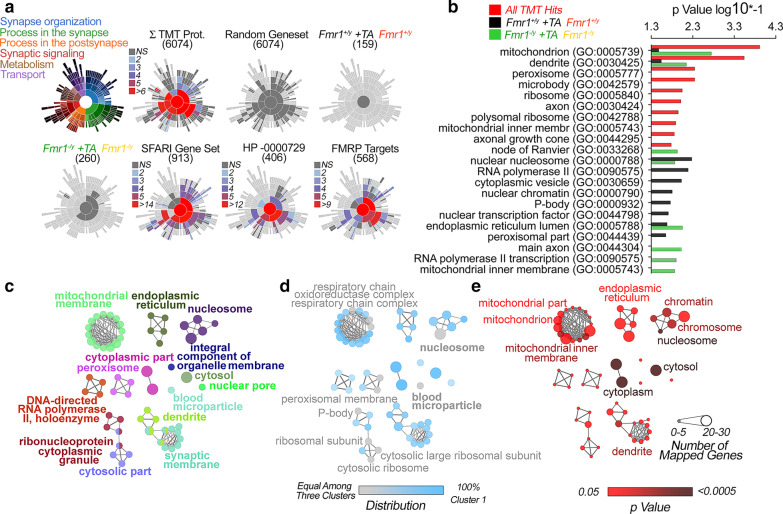
Fig. 3.
Ontological Analysis of the Proteome Sensitive to Activity Deprivation and Fmr1−/y. a Synaptic Ontology Analysis using the SYNGO tool. Sunburst plots represent synaptic annotated GO BP ontologies. Colors represent -log10 FDR corrected p values. Gene sets are defined in Fig. 1. SFARI correspond to the autism spectrum disorder genes curated by https://gene.sfari.org/database/human-gene/. HP-0000729 corresponds to the Human Phenotype Ontology annotated term Autistic behavior HP:0000729 (https://hpo.jax.org/app/browse/term/HP:0000729). b GO CC Term ontology analysis performed with the ENRICHR tool. All proteins sensitive to TTX-APV plus those sensitive to the Fmr1−/y genotype (red bars) were compared to wild type TTX-APV treated hits (black bars) and Fmr1−/y TTX-APV treated hits (green bars). c ClueGO Ontology Analysis of all TMT Hits defined by the red bar in B. Node size represents number of mapped genes. d ClueGO Ontology gene percentage contributions among the three gene sets in b. e ClueGO Ontology Statistics. Non-corrected Two Sided Hypergeometric Test. See Additional file 2: Table S2