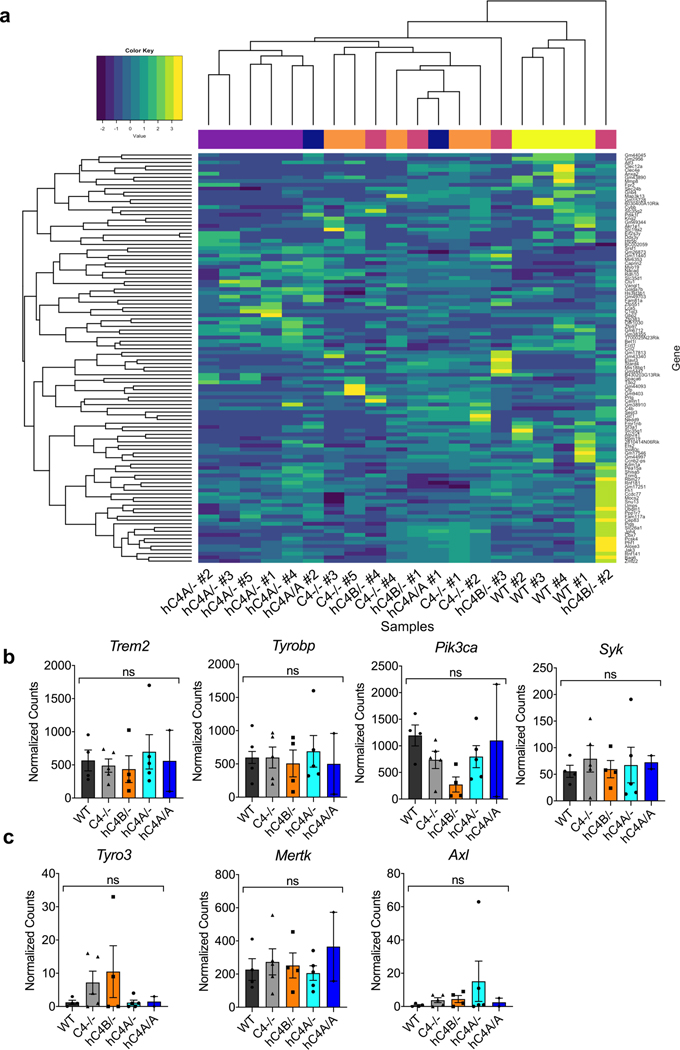
Extended Data Fig. 8 |. Microglial RNA sequencing analysis reveals no transcriptomic alterations in hC4 transgenic mice.
a, Bulk RNA sequencing analysis of microglia isolated from the frontal cortex of adolescent (P40) mice from WT (n = 4), C4−/− (n = 5), and hC4A/- (n = 5), hC4B/- (n = 4), and hC4A/A (n = 2) groups. Heatmap representation of differentially expressed genes between all experimental groups shows no significant transcriptional profile difference between any two groups. b-c, Normalized gene counts for the TREM2/DAP12 signaling pathway (b) and TAM receptor genes (c) were calculated for WT (n = 4), C4−/− (n = 5), hC4A/- (n = 5), hC4B/- (n = 4), and hC4A/A (n = 2; ns = P > 0.05, Kruskal-Wallis test with Dunn’s test). Bar graphs show mean ± s.e.m.