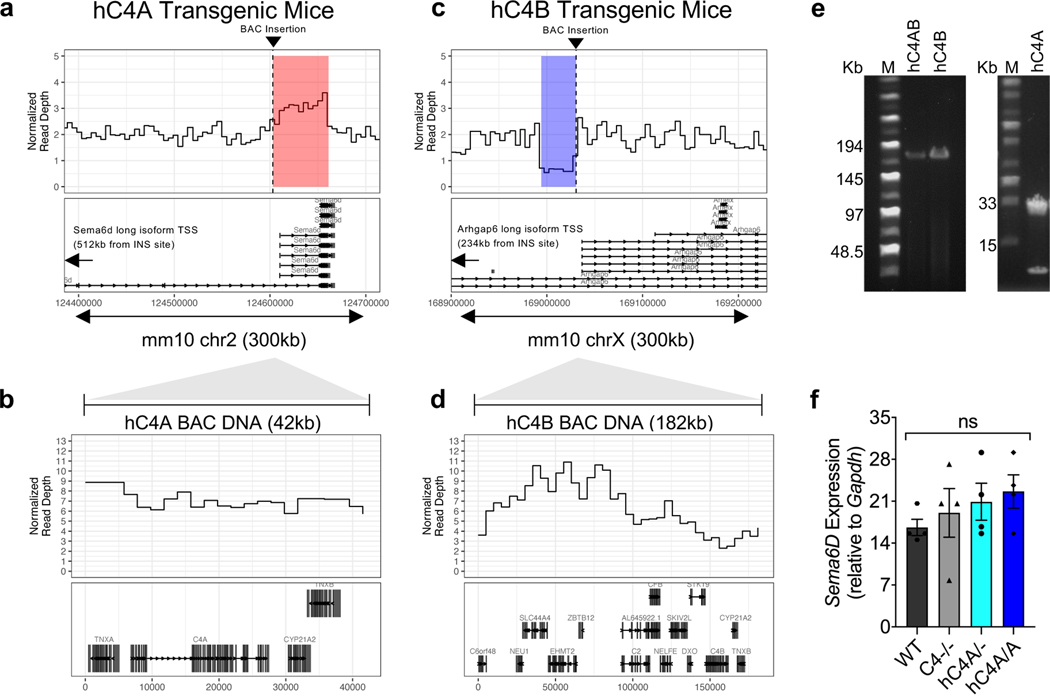
Extended Data Fig. 1 |. Characteristics of C4 BAC DNA integration.
a-d, Insertion sites and associated rearrangements are shown for each mouse strain. a, c, The BAC DNA insertion site is shown for each mouse strain. Normalized read depth in 2-kb genomic windows at the insertion sites of the BACs. Dashed vertical lines indicate the insertion site. The hC4A insertion was associated with a duplication, and the hC4B insertion was associated with a small deletion. b, d, The BAC DNA-associated rearrangements are shown for each mouse strain. Normalized read depth in 1-kb genomic windows shows the copy number of the inserted constructs. Gene models are shown beneath the read depth plots. Human C4A was likely inserted into hC4A transgenic mice at a normalized read depth between 5 and 9. Human C4B was likely inserted into hC4B transgenic mice at a copy number between 2 and 5. The inserted hC4B BAC constructs contain additional internal copy number variants. e, Pulse-field gel showing linearized hC4B and hC4A BAC DNA used for microinjection into zygotes (representative of 3 independent experiments). Unprocessed versions of the gels can be found in Supplementary Figure 1a. f, Sema6D mRNA expression level in cortex was not changed by the region duplication caused by hC4A BAC DNA insertion. Gapdh was used as a control housekeeping gene (n = 4 mice per group; ns = P > 0.05, Kruskal-Wallis test with Dunn’s multiple comparisons test). Bar graph shows mean ± s.e.m.