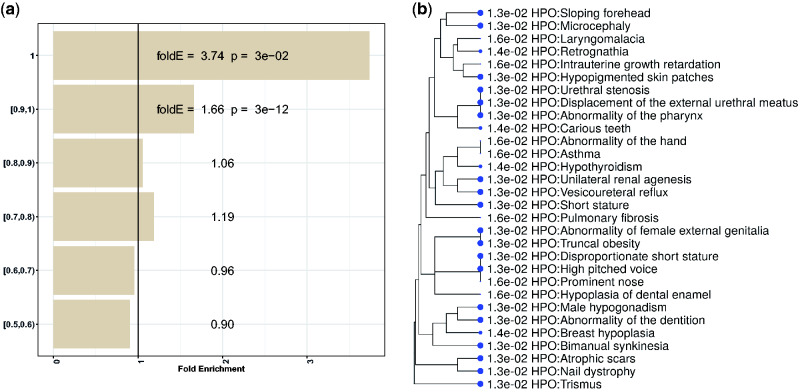
Fig. 1.
(a) Fold enrichment of variants for coding exonic sequences in different FST bins (y axis). (b) Human phenotypes that are enriched for genes carrying at least one missense exonic variant with an FST value greater than or equal to 0.5. All P-values are FDR-corrected and FDR threshold 0.05 is used. The size of the blue dots is proportional to the FDR. The human phenotype ontology database (https://hpo.jax.org/app/) was used for enrichment analyses due to the lack of such functional annotation for the canine genes. The tree is constructed using the distance between two gene sets based on the number of genes in the intersection and the union of two sets. The distance matrix is then used to construct a hierarchical clustering tree based on the number of shared and unique genes between the different sets.