Figure 1.
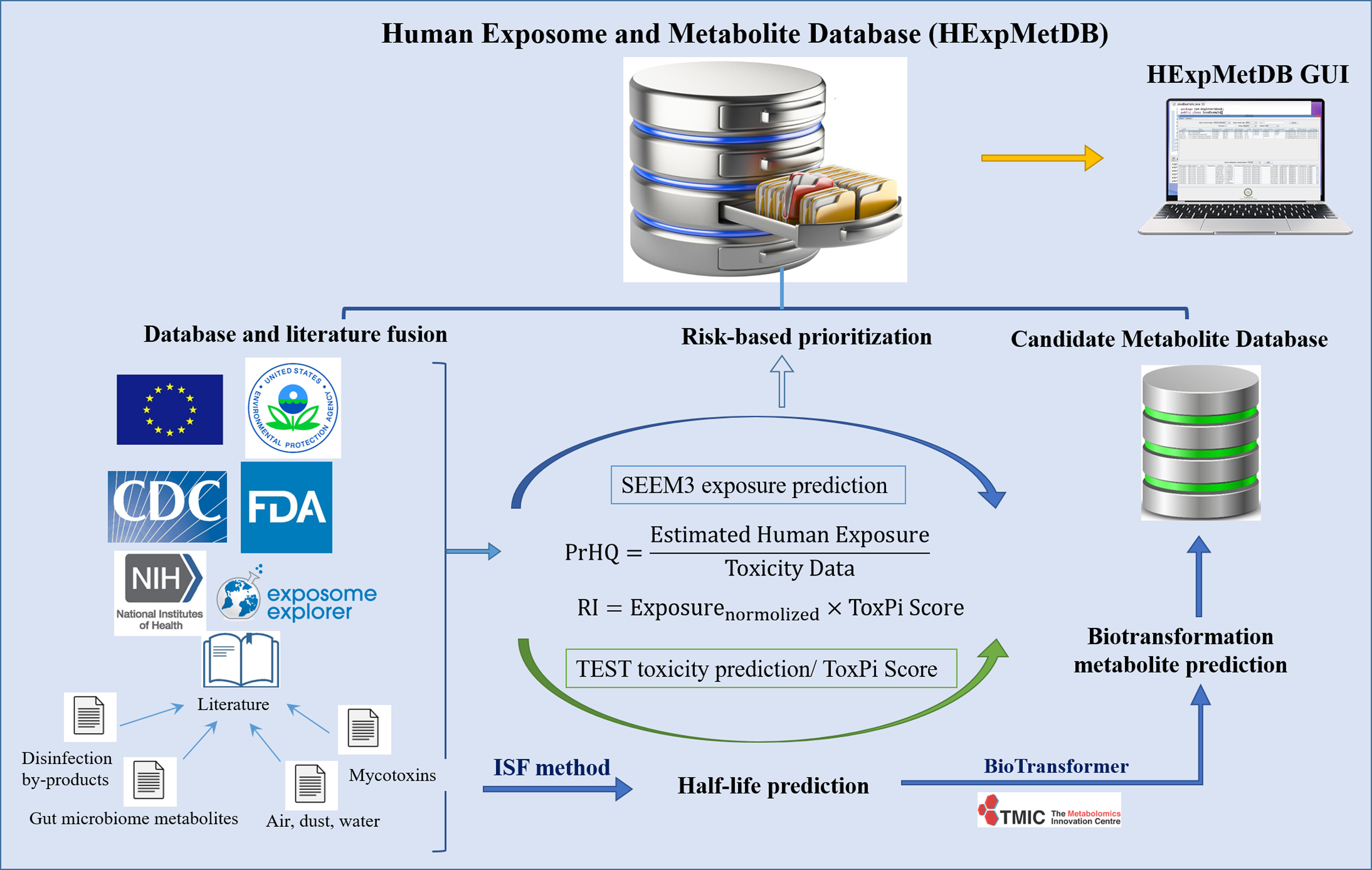
Schematic workflow for Human Exposome and Metabolite Database (HExpMetDB) establishment. Note: CDC, Centers for Disease Control and Prevention; FDA, U.S. Food and Drug Administration; GUI, graphical user interface; ISF, Iterative Fragment Selection; NIH, National Institutes of Health; PrHQ, probabilistic hazard quotient; RI, risk index; SEEM3, Systematic Empirical Evaluation of Models; TEST, Toxicity Estimation Software Tool; ToxPi, Toxicological Priority Index.
