Figure 1.
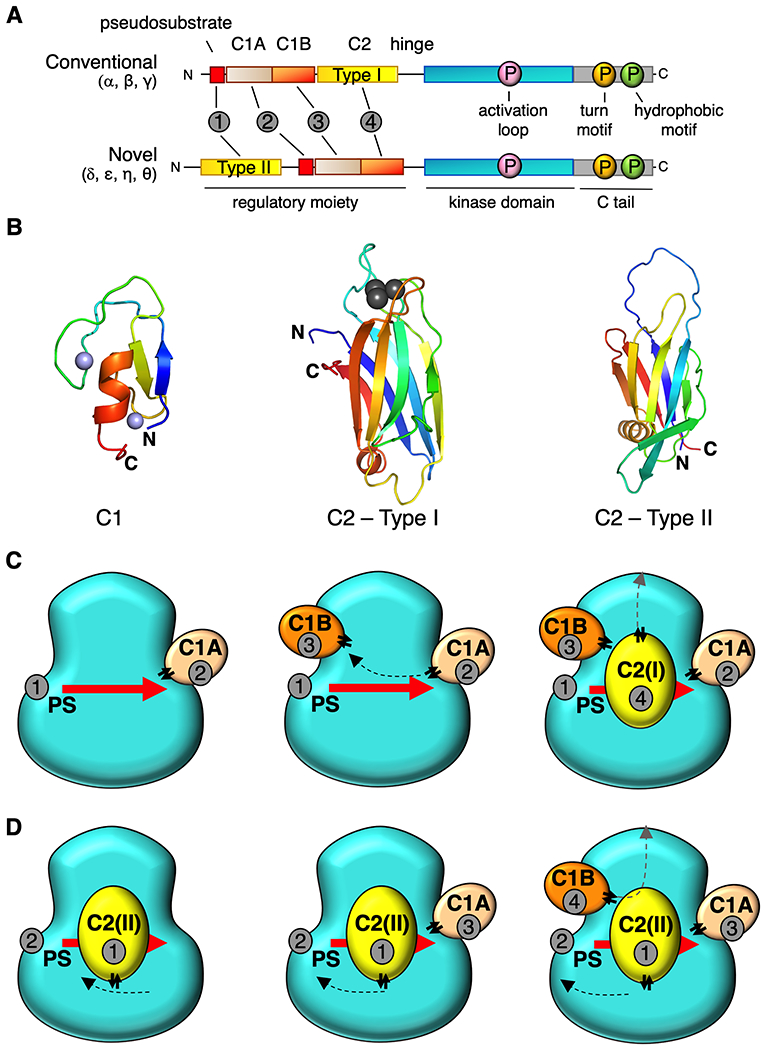
Proposed architecture for conventional and novel PKC isozymes. (A) Primary structure and domain composition of conventional and novel PKC isoforms showing pseudosubstrate (PS, red), C1A domain (sand), C1B domain (orange), C2 domain (yellow), kinase (cyan), and C-tail (grey). Three processing phosphorylations indicated in magenta, orange, and green for the activation loop, turn motif, and hydrophobic motif, respectively. Order of domains indicated by grey circles. (B) Structure of C1 domain (PKCδ, PDB ID code 2YUU)) and two types of C2 domain: Type1 C2 (PKCβ PDB ID code 1A25) in conventional PKCs and Type 2 C2 (PKCθ PDB ID code 2ENJ) in novel PKCs highlighting different topology but same overall architecture. Notably, the N and C termini are on opposite ends of each the two types of C2. Zn2+ ions (C1 domains) in purple and Ca2+ ions (Type I C2 domain) in grey (C) Proposed positions of each domain on the kinase module for conventional PKCs and novel PKCs (D) showing common tertiary structure. Domains are numbered in the order they appear in the primary structure in (A). Double arrows indicate positions of the “plugs”.
