Figure 2.
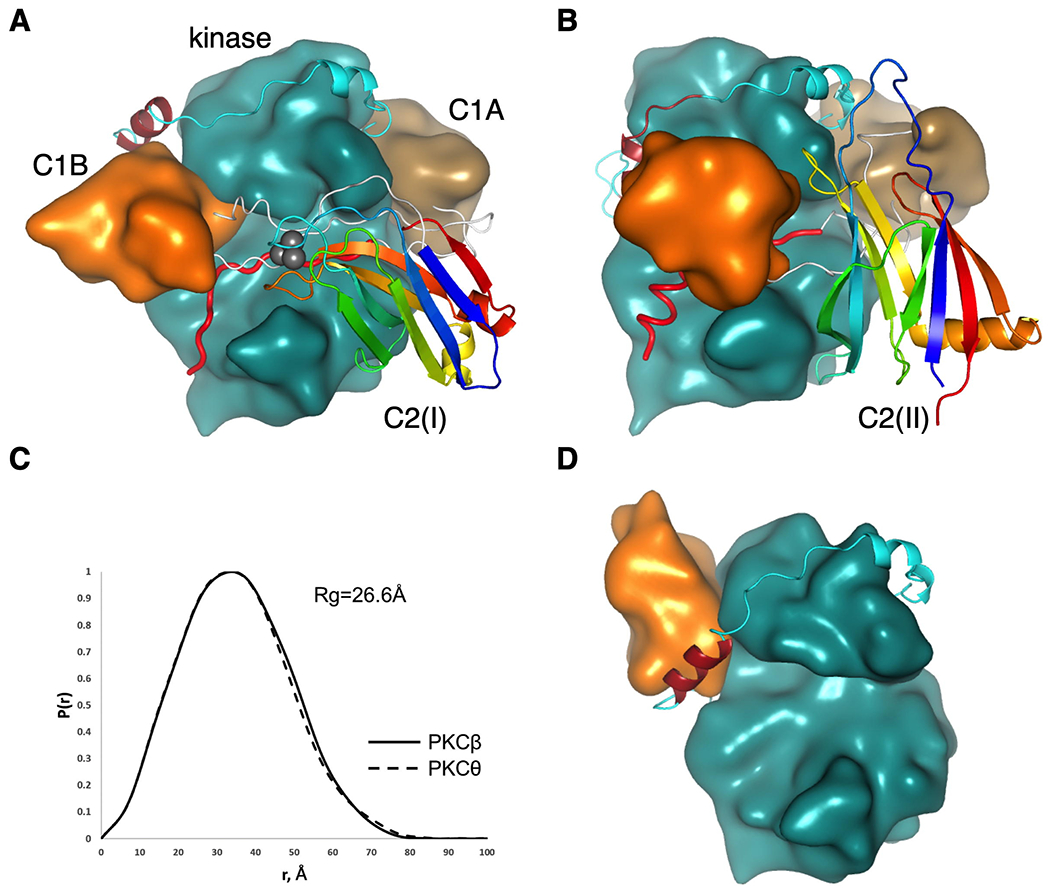
Conserved architecture of conventional and novel PKC isozymes. (A) Proposed structure of PKCβII highlighting positions of the C1B (orange), NFD helix (burgundy) in the kinase domain (cyan), C1A domain (sand) and C2 domain (ribbon diagram colored from blue (N terminus) to red (C terminus) to indicate topology; Ca2+ ions in grey). Linkers connecting domains are shown in white with the pseudosubstrate (red) positioned in the active site. (B) Proposed structure of PKCθ, colored as in (A) showing inverted C2-domain topology yet same overall structure. (C) Pair distance distribution function P(r) computed from the proposed models showing similar shapes and radii of gyration of conventional PKCβ and novel PKCθ. (D) Previously reported structure of PKCβII (12) (pdb:3PFQ) showing C1B domain (orange) positioned on top of NFD helix (burgundy).
