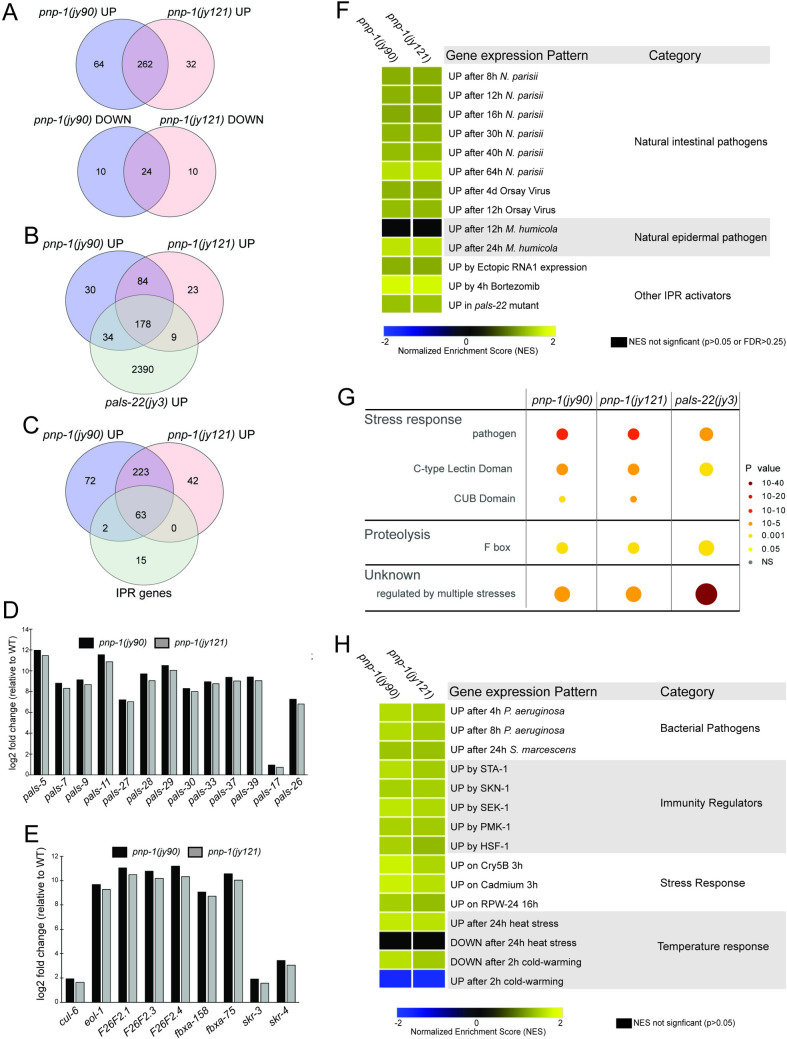
Fig 5. RNA-seq analysis demonstrates that pnp-1 represses expression of many IPR genes.
A) Venn diagram of differentially expressed genes in pnp-1(jy90) and pnp-1(jy121) mutants as compared to wild-type animals. Both up (rf = 31.5; p < 0.000e+00) and down (rf = 239.6; p < 3.489e-58) regulated genes have significant overlap between the two mutant alleles. B) Venn diagram of upregulated genes in pnp-1(jy90), pnp-1(jy121) and pals-22(jy3) mutants as compared to wild-type animals. Upregulated genes in both pnp-1(jy90) (rf = 2.9; p < 4.566e-62) and pnp-1(jy121) (rf = 2.8; p < 1.591e-52) mutants have significant overlap with those upregulated in pals-22(jy3) mutants from a previous study [17]. C) Venn diagram of upregulated genes in pnp-1(jy90), pnp-1(jy121) and the IPR genes (defined in [17]). Upregulated genes in both pnp-1(jy90) (rf = 28.8; p < 1.242e-88) and pnp-1(jy121) (rf = 30.9; p < 2.567e-87) mutants have significant with the IPR genes [17]. A-C) rf is the ratio of actual overlap to expected overlap where rf > 1 indicates overrepresentation and rf < 1 indicates underrepresentation (see S7 Table for more detail) D) Log2 fold-change of a subset of pals genes in pnp-1(jy90) and pnp-1(jy121) mutants normalized to wild-type. E) Log2 fold change of a subset of non-pals genes in pnp-1(jy90) and pnp-1(jy121) mutants normalized to wild-type. F) Correlation between genes differentially expressed by various known IPR activators and those differentially expressed in pnp-1(jy90) or pnp-1(jy121) mutants. G) Wormcat analysis for significantly enriched categories in differentially expressed gene sets of pnp-1(jy90) and pnp-1(jy121) mutants. Size of the circles indicates the number of the genes and color indicates value of significant over representation in each Wormcat category. H) Correlation of genes differentially expressed by bacterial pathogens, immune regulators, and various stressors to those differentially expressed in pnp-1(jy90) or pnp-1(jy121) mutants. F, H) Analysis was performed using GSEA 3.0 software, and correlations of genes sets (S5 Table) were quantified as a Normalized Enrichment Score (NES) (S6 Table). NES’s presented in a heat map. Blue indicates significant correlation of downregulated genes in a pnp-1 mutant with the tested gene set, yellow indicates significant correlation of upregulated genes in a pnp-1 mutant with the tested gene set, while black indicates no significant correlation (p > 0.05 or False Discovery Rate < 0.25).