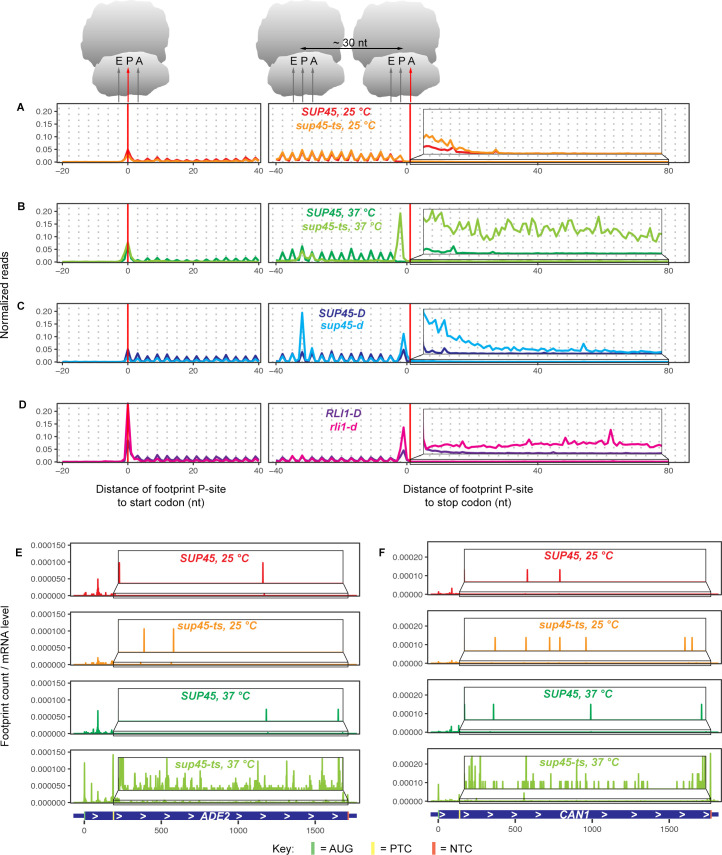
Fig 1. Ribosome occupancy at and beyond the stop codon increases in yeast cells defective in translation termination or ribosome recycling.
A-D. Ribosome footprints (2,693 genes with 3’-UTR annotations and without sequence overlapping) normalized to the total number of footprints in the indicated nt window (-20 to 40 nt around the start codon and -40 to 80 nt around the stop codon) aligned at their start or canonical stop codons. Footprints were plotted by the position of their P-sites. Each panel contains the data from WT and its respective mutant. The elevated number of footprints at the penultimate codon of the ORFs in mutant samples demonstrates ribosome stalling when the stop codon was in the A site. (The footprints at this codon and their shift to reading frame +1 in other yeast strains obtained from published data may be attributable to the differences in strain background or sequencing library preparation procedures.) Inset: Magnified view of the region in the box, showing increased 3’-UTR ribosome occupancy in sup45-ts cells at 37°C, sup45-d, and rli1-d cells relative to their respective WT. E-F Ribosome footprints normalized to the respective mRNA levels mapped across the ORF of two PTC-containing alleles, ade2-1 (E) and can1-100 (F), in SUP45 and sup45-ts strains at 25°C and 37°C.