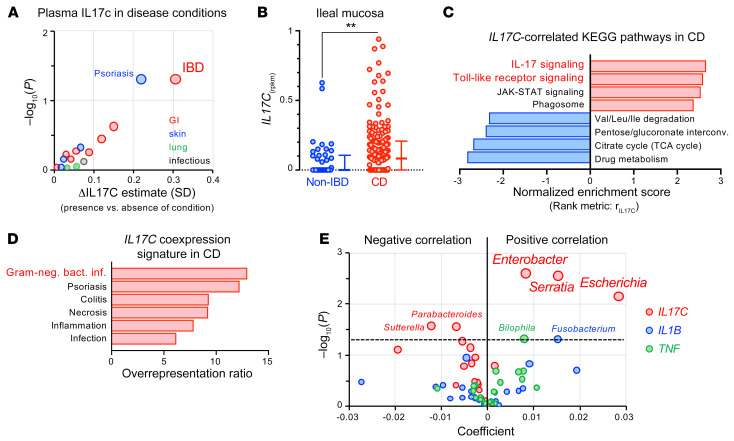
Figure 6. IL17C induction observed in a subset of patients with IBD is a marker for abnormal epithelial stimulation by gram-negative bacteria.
(A) Positive associations of plasma IL-17C concentration with self-reported health history of study participants considering GI, skin, lung, and chronic infectious disease categories. Shown is the average difference in standardized plasma IL-17C for presence versus absence of a condition. We evaluated the nominal significance of effects using the Welch 2-sample test adjusted for age, sex, body mass index, season, and ancestry. See Supplemental Table 11 for detailed results. (B) Expression of IL17C in ileal mucosal biopsies from patients with CD (n = 174) and non-IBD controls (n = 42) from the RISK cohort. Error bars represent medians with IQR. Two-tailed Mann-Whitney. **P = 0.0027. (C) Gene set enrichment analysis using correlation with IL17C expression (rIL17C) as the rank metric to identify IL17C-correlated KEGG pathways in the mucosal biopsies of CD patients (FDR < 0.05). See Supplemental Tables 13 and 14 for additional information. (D) Overrepresentation of IL17C-coexpression signature (rIL17C > 0.5) in disease-associated gene sets from the GLAD4U database (59) (FDR < 0.05; Supplemental Table 15). (E) Multivariate association analysis using the expression of IL17C and proinflammatory cytokines (TNF, IL1B) in ileal CD biopsies (n = 135) as predictors and genus-level microbial abundance data of the mucosal microbiome as a response. Positive coefficients indicate a positive correlation between gene expression and compositional abundance of a bacterial genus (see Supplemental Tables 16 and 17 for input data and detailed results).