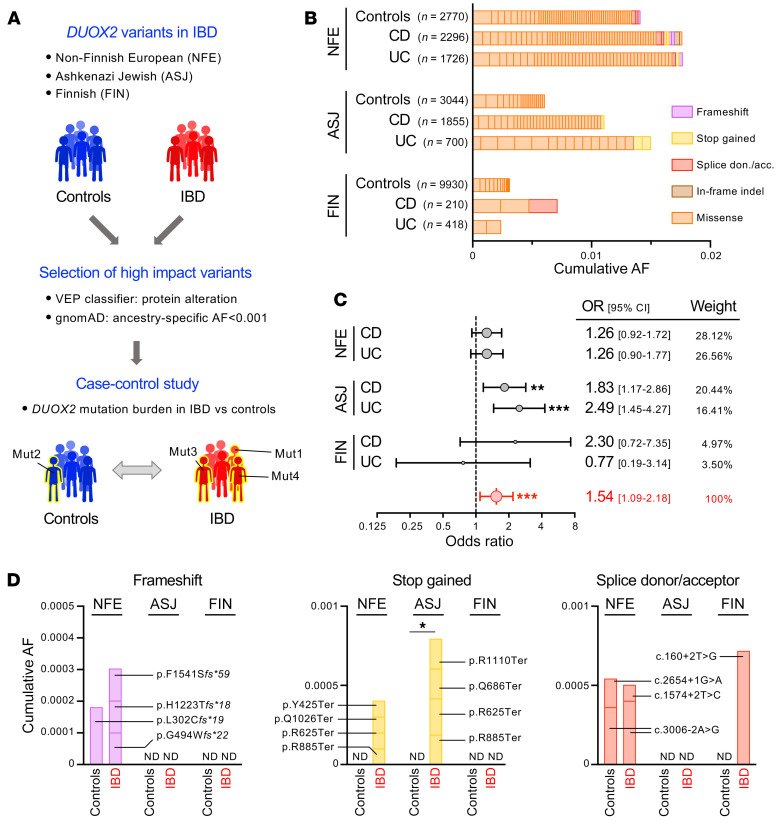
Figure 7. High impact DUOX2 variants confer increased risk for IBD.
(A) Outline of the case-control study comparing the burden of high-impact DUOX2 protein variants in patients with IBD and ancestry-matched non-IBD control cohorts. We stratified variants using population-specific allele frequencies from the gnomAD database. (B) Contribution of individual high impact DUOX2 protein variants to the cumulative allele frequencies. NFE, non–Finnish European; ASJ, Ashkenazi Jewish; FIN, Finnish. Note that the low prevalence of very rare variant carriers in Finnish participants is due to multiple genetic bottlenecks in that isolated population (23). See Supplemental Tables 18–20 for detailed data and Supplemental Figure 6 for the distribution of variants with higher allele frequencies. (C) Carriers of high-impact DUOX2 protein variants are at increased risk for developing IBD. The Forest plot depicts estimated ORs with 95% CI for patients with UC and CD from the 3 ancestry cohorts. The combined OR was calculated using a random-effects model with the Mantel-Haenszel weighting method (Supplemental Table 21). Test of the null hypothesis that OR is equal to 1 (60). (D) Detailed view of DUOX2 variants with predicted complete loss-of-function (i.e., frameshift, stop gained, and splice donor or acceptor site variants) in IBD and control cohorts. Two-tailed Fisher’s exact test. ND, not detected. *P < 0.05; **P < 0.01; ***P < 0.001.