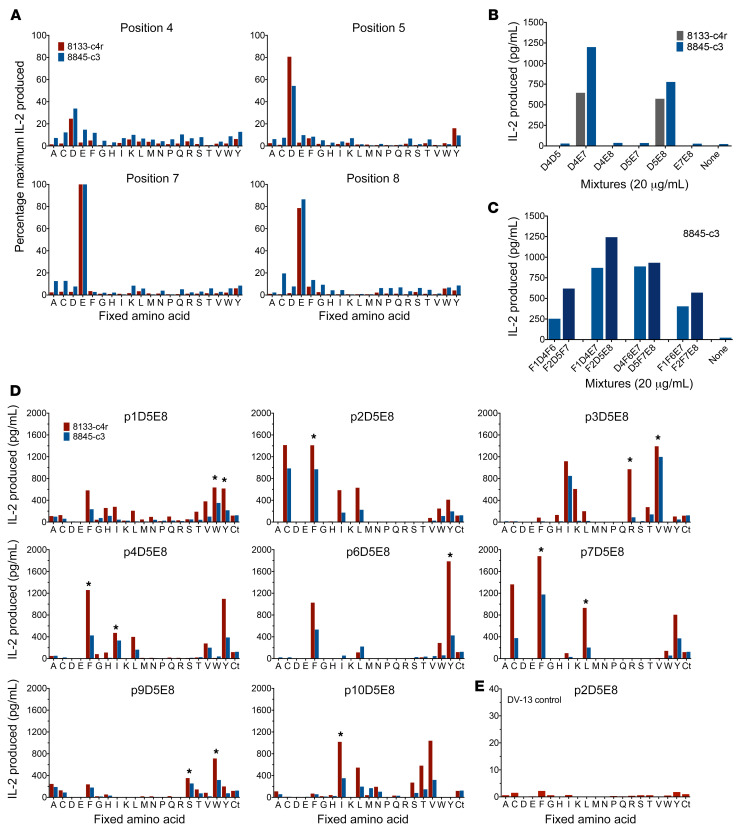
Figure 2. Antigen discovery of BAL-derived LKGGG CDR3β TCRs using unbiased and biased decapeptide PSLs.
(A) Representative results of 2 experiments depicting IL-2 responses (pg/mL, done in duplicate) of hybridomas 8133-c4r and 8845-c3 to an unbiased PSL. Peptide mixtures (200 μg/mL) were presented by HLA-DP2–transfected fibroblast line DP2.21 in the presence of BeSO4 (75 μM), and data were normalized to peak IL-2 produced in the assay for each hybridoma (mixture E7, lower left). Label on x axis denotes amino acid (single letter code) fixed at each defined position. Four (p4, p5, p7, p8) of 10 peptide positions scanned are displayed. (B and C) IL-2 produced (pg/mL) by indicated hybridomas in response to select dual-defined (100 μg/mL) (B) and triple-defined (20 μg/mL) (C) mixtures in the presence of BeSO4 (75 μM). (D and E) IL-2 responses (pg/mL) of hybridomas 8133-c4r and 8845-c3 (D) and hybridoma DV-13 (E) to a biased PSL with 2 positions fixed (D5E8). Each panel shows a different peptide position scanned using peptide mixtures (20 μg/mL) tested in duplicate in the presence of BeSO4 and including a control D5E8 mixture (Ct). Asterisks indicate amino acids selected at each peptide position for synthesis of mimotopes.