Fig. 5. SE-driven lncRNAs in GBM.
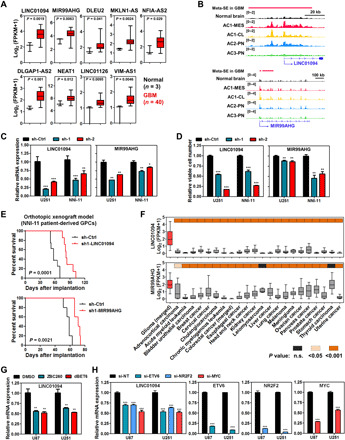
(A) Elevated expression of SE-driven lncRNAs in GBM samples (cohort 1). Student’s t test was applied. (B) Differential deposition of H3K27ac signals across regulatory regions of LINC01094 and MIR99AHG in GBM subtypes and normal brain tissues. (C to E) Effect of shRNA-mediated silencing of LINC01094 and MIR99AHG on target expression (C) and cell viability (D), and survival of recipient mice with intracranial transplantation of NNI-11 cells (E). (F) Differential expression of LINC01094 and MIR99AHG transcripts among various human cancers. (G) Effect of BET protein degraders on the expression of LINC01094. U87 and U251 GBM cells were treated with either ZBC260 or dBET6 (200 nM, 8 hours) before harvest. (H) Effect of siRNA-mediated silencing of core TFs on the expression of LINC01094. GBM cells were transfected with indicated siRNAs (48 hours) and subjected to qPCR analysis. Either one-way ANOVA or t test was used for analysis of significance. Error bars in (C), (D), (G), and (H) represent SEM, n = 3. Log-rank test was applied for survival analysis in (E). Boxplots in (A) and (F) represent the 25th and 75th percentiles, with midlines indicating the median values and whiskers extended to the lowest/highest values. n.s., not significant.
