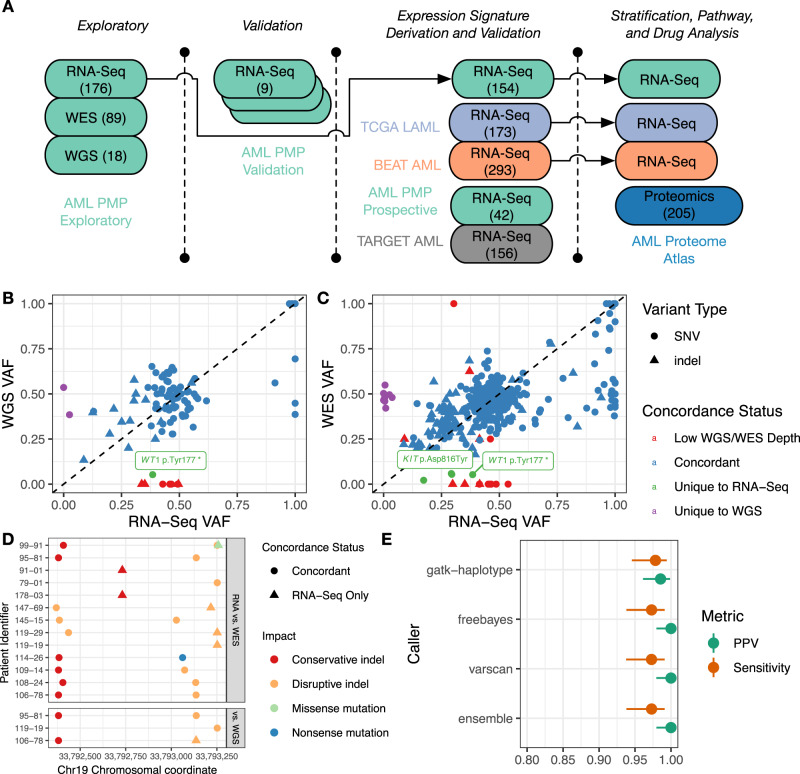
Fig. 1. Experimental overview and short nucleotide variant/indel analysis.
A Overview of datasets used at each stage of the analysis, colored by sequencing project, with sample size for each data set indicated in brackets. For the AML PMP exploratory batch, MDS samples were used for profiling the SNV and gene fusion pipelines, but not for expression-based analyses, and validation RNA-Seq libraries were prepared in triplicate. B–C Matched variant allele frequency (VAF) for variants between whole-genome sequencing (WGS) and RNA-Seq (B) and whole exome sequencing (WES) and RNA-Seq (C), by concordance status, variant type, and coverage status (sites with ≤10x coverage are indicated as ‘Low WGS/WES Depth’). Selected variants discussed in the text are labeled with Human Genome Variation Society (HGVS) nomenclature. D Variant observations in CEBPA. Potentially disruptive mutations for each patient (y-axis) are indicated by their chromosomal coordinate (x-axis), with predicted impact and variant concordance. E Sensitivity and positive predictive value (PPV) for each variant caller in the validation cohort (with 95% confidence intervals).