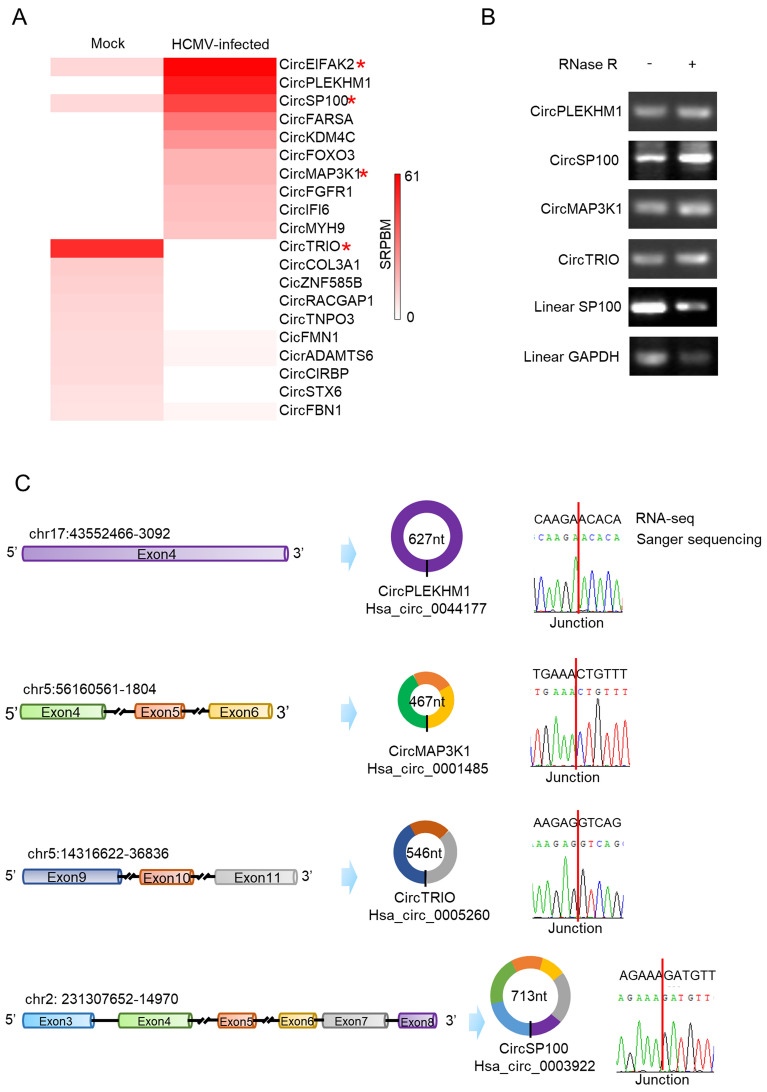
Fig. 3.
Validation of candidate circRNA transcriptions changed by HCMV productive infection. A Heatmap of the significant up-regulated and down-regulated circRNAs. Red star symbols mark the circRNAs selected for further verification; B Verification of cyclization for the selected circRNAs by RNase R treatment. RNase R treatment indicates noticeable depletion of linear RNA (SP100 and GAPDH) but not affect the circRNAs (circPLEKHM1, circSP100, circMAP3K1, and circTRIO); C Verification of the selected circRNAs by Sanger sequencing. Sanger sequencing indicated all of the chosen circRNAs were consistent with RNA-seq results.