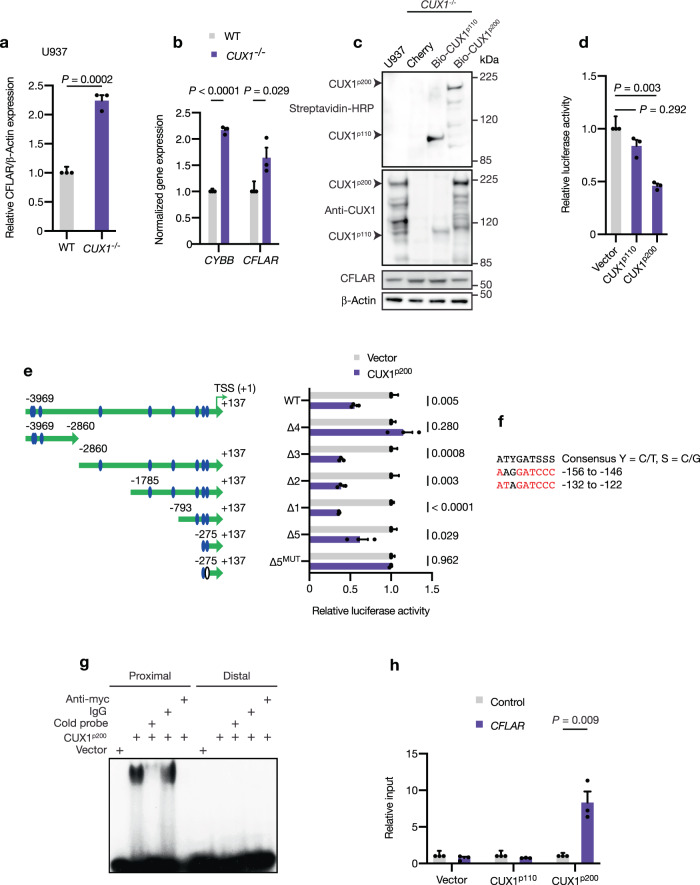
Fig. 7. CFLAR is directly repressed by CUX1.
a Quantification of CFLAR protein levels in CUX1−/− U937 cells compared with wild-type. β-Actin was used as a loading control. The experiment was performed three times. b Expression of CYBB and CFLAR transcripts normalized to HPRT1 in CUX1−/− U937 cells compared with wild-type by qRT-PCR. Biological and technical triplicates were analyzed. c Immunoblot showing expression of biotinylated CUX1p100 and CUX1p200 proteins (top) in doxycycline-treated CUX1−/− U937 cells harboring the indicated constructs. The corresponding levels of CFLAR are also shown (bottom). β-Actin was used as a loading control. The experiment was performed three times. d Promoter-luciferase assays showing luciferase activity in HEK293T cells transfected with the indicated constructs and a ~4 kb CFLAR promoter-luciferase reporter. Values were normalized to empty vector. Three experiments were performed with triplicate samples in each experiment. e Promoter-luciferase assays with deletion constructs of the CFLAR promoter-luciferase reporter (large green arrow). Blue ovals, potential CUX1-binding sites. White oval, mutated potential CUX1-binding site. TSS, transcriptional start site (nominated + 1 position). Three experiments were performed with triplicate samples in each experiment. f Sequence of potential CUX1-binding sites in CFLAR promoter closest to TSS. Locations of binding sites relative to TSS are shown. Consensus CUX1-binding site is shown (top). Red, exact consensus match. g Representative electrophoretic mobility shift assay with indicated components and proximal (−132 to −122) or distal (−156 to −146) radiolabeled, potential CUX1-binding dsDNA probes. The experiment was performed twice with similar results. h Chromatin immunoprecipitation assay followed by qRT-PCR for the CFLAR promoter region (purple) or distal control region (gray) in CUX1−/− U937 cells complemented with the indicated constructs as shown in c. Biological and technical duplicates were analyzed. All plots show mean + s.e.m. Two-tailed, unpaired t-test.