Abstract
Objective(s):
Doxorubicin (Dox) is one of the most well-known chemotherapeutics that are commonly applied for a wide range of cancer treatments. However, in most cases, efflux pumps like P-glycoprotein (P-gp), expel the taken drugs out of the cell and decrease the Dox bioavailability. Expression of P-gp is associated with elevated mRNA expression of the ATP-binding cassette B1 (ABCB1) gene.
Materials and Methods:
In the current study, different sequences of cell-penetrating peptides (CPPs) containing tryptophan, lysine, and arginine and their nano-complexes were synthesized and their impact on the expression and activity of the ABCB1 gene was evaluated in the A549 lung carcinoma cell line. Furthermore, the cellular uptake of designed CPPs in the A549 cell line was assessed.
Results:
The designed peptides, including [W4K4], [WR]3-QGR, R10, and K10 increased Dox cytotoxicity after 48 hr. Furthermore, arginine-rich peptides showed higher cellular uptake. Rhodamin123 accumulation studies illustrated that all the obtained peptides could successfully inhibit the P-gp pump. The designed peptides inhibited the ABCB1 gene expression, of which, [W4K4] resulted in the lowest expression ratio.
Conclusion:
[W4K4], [WR]3-QGR, R10, and K10 could successfully increase the Dox cytotoxicity by decreasing the efflux pump gene expression.
Key Words: Cancer therapy, CPPs, Doxorubicin, Multi-drug resistance, P-gp
Introduction
Cancer, uncontrolled growth of abnormal cells, is considered a global crisis for the health community (1). In this regard, there has been a growing concern upon applying novel diagnostics and therapeutics to reduce its mortality (2, 3). Lung cancer is known as one of the most commonly occurring cancers in adults that leads to death in most cases (4). Although chemotherapy is an indispensable part of cancer treatment (5), two main impediments reduce its efficiency. First, multidrug resistance (MDR) which is the leading cause of most chemotherapy failures and second is the destructive side effects of chemotherapeutics as a result of their unselective entrance to both normal and tumor cells (6). MDR is a cellular hindrance towards chemotherapy in which tumor cells do not respond to therapeutic agents (7). A wide range of mechanisms is involved in MDR ranging from cellular factors to physiological conditions. In another view, this drug resistance could be based on cancer cell genetics and gradual response to chemotherapeutics over the period of treatment, which are called inherent and acquired responses, respectively (8, 9). Mild to harsh acidic environment, hypoxia, abnormal vascularization, as well as high interstitial fluid pressure are some of the physiological conditions related to MDR (10, 11). On the other hand, drug resistance occurs due to numerous cellular factors, including enhanced drug metabolism and detoxification, reduced bioactivation of drugs, disruption of apoptotic signaling pathways, and reduced influx of drugs, namely increased drug efflux (11-16). Activation of special efflux pumps (MDR proteins) such as P-glycoprotein (P-gp) across the membrane is recognized as a crucial cause for MDR (17-20). ATP-binding cassette (ABC) is one of the most studied MDR protein superfamilies that is physiologically responsible for transporting different substrates via special pumps across the membrane by ATP consumption against their concentration gradient (19, 21, 22). There are 48 genes related to ABC proteins which are mainly expressed in human cells. Based on their functions and structural similarities, ABC proteins are divided into seven subfamilies (ABC-A to ABC-G), three of which have strongly been approved to be responsible for MDR including MDR-associated protein (MRP1, ABCC1), breast cancer resistance protein (MXR or BCRP, ABCG2), and P-gp (MDR1, ABCB1) (23-25). P-gp is a single polypeptide comprising twelve transmembrane domains (TMDs) and two nucleotide-binding domains (NBDs) (26) that transports a wide range of compounds such as anticancer drugs (Doxorubicin, Paclitaxel, Vincristine, Bisantrene, etc.), HIV protease inhibitors (Ritonavir, Saquinavir, Nelfinavir), analgesics (Morphine), antihistamines (Terfenadine, Fexofenadine), natural products (Curcuminoids, Colchicine), antibiotics (Erythromycin, Gramicidin A), lipids, and peptides (24). P-gp is mainly expressed in all cells but is overexpressed in cancer cells (17). Doxorubicin (Dox, Adriamycin) is an antineoplastic or cytotoxic chemotherapeutic which is applied for treatment of different cancers. However, due to the robust drug resistance provided by cancer cells against Dox, its therapeutic efficacy is significantly limited (27). In the recent decade, a wide range of approaches like silencing MDR-related miRNAs, altering regimen of chemotherapeutics (combination therapy), applying monoclonal antibodies reversing P-gp, designing novel agents that are not recognized by P-gp, P-gp modulators, and innovative classes of nanometric drug delivery systems (DDSs) have been introduced and investigated to tackle drug resistance and improve chemotherapy efficiency (7, 17, 28, 29). It has been proposed that amphiphilic, cationic molecules could inhibit negatively charged P-gp efflux pump (30). Therefore Cell-penetrating peptides (CPPs) have gained much attention due to their ability to overcome P-gp related drug resistance. CPPs are nano-scale short sequences of amino acids with cationic or amphiphilic features (31, 32). CPPs are able to deliver different kinds of cargoes into various cells in a nontoxic manner (33, 34). In this study, for the first time, ABCB1 expression and P-gp activity on Dox-resistant A549 lung cancer cell line were investigated in the presence of amphiphilic-cationic CPPs and their nano-complexes.
Materials and Methods
Materials
O-(Benzotriazole-1-yl)-N,N,N,N′-tetramethyluronium tetrafluoroborate (TBTU), hydroxybenzotriazole (HOBt), trifluoroacetic acid (TFA), 5(6)-carboxyfluorescein (FAM), triisopropylsilane (TIPS), phenol, piperidine, ethanol (95%), isopropanol, trizol, and chloroform were obtained from Sigma-Aldrich (Sigma, Saint, Louis, USA). N-ethyldiisopropylamine (DIPEA) was acquired from Merck (Merck, Germany). Dimethylformamide (DMF) and dichloromethane (DCM) were purchased from Scharlau Chemie (Barcelona, Spain). Fluorenylmethyloxycarbonyl (Fmoc)-amino acid derivatives and 2-chlorotrityl chloride resin (capacity: 1.08 mmol g−1) were purchased from AAPPTec (Louisville, KY, USA). RPMI-1640 medium and fetal bovine serum (FBS) were purchased from Gibco (Grand Island, NY, USA), Penicillin/streptomycin solution was provided by Applichem (Darmstadt, Germany). 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) was obtained from Roth (Roth, Germany). Trypsin-EDTA (ethylenediaminetetraacetic acid) was obtained from Invitrogen (Invitrogen, Carlsbad, USA). Cell culture T-75 flasks and 6- and 96-well plates were obtained from Biofil (Canada) and SORFA (China). A549 cell line was provided by the Pasteur Institute (Tehran, Iran).
Synthesis of CPP
Peptides were manually synthesized by the solid-phase peptide synthesis (SPPS) method on 2-chlorotrityl chloride resin in glass reaction vessels (35, 36). Eleven different sequences were synthesized, and their nano-complexes were prepared (Table 2). Briefly, CPPs were synthesized by swelling 250 mg of resin in 3 ml dry DCM for 1 hr under dry nitrogen. Then, the DCM was drained from resin and Fmoc-protected amino acid was dissolved in dry DMF and poured into the reaction vessel, and shaken to be coupled to the resin in the presence of DIPEA for almost three hours. The coupling process was monitored by the ninhydrin test. In this step, other reacting trityl groups were end-capped using 0.2 ml ethanol and shacking for 15 min. The ethanol was filtered and washed by DCM (2×2 ml), DMF (2×2 ml), and methanol (2×2 ml). After coupling of the first amino acid, its FMOC group was deprotected by adding piperidine/DMF solution (20% v/v, 2 ml) and shaking for 30 min under argon atmosphere. The Ninhydrin test was carried out to monitor the deprotection process. Other amino acids were also coupled to the peptide chains in the presence of appropriate amounts of TBTU, HOBt, and DIPEA and in a repeated cycle of deprotection, washing, coupling, and washing. Peptide derivatives were cleaved and deprotected by adding 10 ml of cleavage cocktail (TFA/H2O/Phenol/TIPS: 88/5/5/2 v/v/v/v). The reaction vessel was then stirred for three hours. Then the volume of filtrates was reduced to one-third of the initial volume using a rotary-evaporator. The peptides precipitated by addition of cold diethyl ether (10 times more than the cocktail volume). Finally, the peptides were centrifuged at 1792 g for 4 min, separated, dried, and stored in Eppendorf tubes at -20 °C.
Table 2.
The sequence and molecular properties of synthesized peptides
| Peptide | C-Sequence-N | Chemical formula | Molecular weight (g/mol) | Isoelectric point | Net Charge at pH=7 | Extinction coefficient M -1 C -1 |
Approximately
volume A 3 |
|---|---|---|---|---|---|---|---|
| P1 | COOH-[WR]4-NH2 | C68H90N24O9 | 1387.62 | pH 12.88 | 4 | 22760 | 1679 |
| P2 | COOH-[WK]4-NH2 | C68H90N16O9 | 1275.56 | pH 11.01 | 4 | 22760 | 1544 |
| P3 | COOH-[W4K4]-NH2 | C68H90N16O9 | 1275.56 | pH 11.01 | 4 | 22760 | 1544 |
| P4 | COOH-[W4R4]-NH2 | C68H90N24O9 | 1387.62 | pH 12.88 | 4 | 22760 | 1679 |
| P5 | COOH-[WR]3-QGR-NH2 | C64H91N25O11 | 1386.59 | pH 12.88 | 4 | 17070 | 1678 |
| P6 | COOH-QGR-[WK]3-NH2 | C64H91N19O11 | 1302.55 | pH 11.73 | 4 | 17070 | 1577 |
| P7 | COOH-R10-NH2 | C60H122N40O11 | 1579.9 | pH 13.35 | 10 | 0 | 1912 |
| P8 | COOH-K10-NH2 | C60H122N20O11 | 1299.76 | pH 11.46 | 10 | 0 | 1573 |
| P9 | COOH-QGR-[WR]3-NH2 | C64H91N25O11 | 1386.59 | pH 12.88 | 4 | 17070 | 1678 |
| P10 | COOH-WRWQGRWRW-NH2 | C69H89N23O11 | 1416.61 | pH 12.7 | 3 | 22760 | 1715 |
| E 10 | COOH-E10-NH2 | C50H72N10O31 | 1309.17 | pH 2.73 | -10 | 0 | 1584 |
** Note: [E: Glutamic Acid, W: Tryptophan, R: Arginine, Q: Glutamine, G: Glycine, K: lysine]
Preparation of CPP/E 10 nano-complexes
Nano-complexes were prepared in the presence of positively charged amino acids of synthesized peptides and negatively charged E10. Peptides were dissolved in dimethyl sulfoxide (DMSO) and E10 was then added in 1:5 ratio (E10: CPP) to form the nano-complexes.
Characterization techniques
The mean hydrodynamic diameter and size distribution of the chosen peptide (P3) and its nano-complexes (P3/E10) were evaluated by a Nano S Zetasizer (Malvern Instruments, Malvern, UK). The sample was diluted in distilled water at pH = 7.4 and the analyses were performed at 25 °C and repeated three times per sample. Scanning Electron Microscopy (SEM) was applied to determine the surface morphology, average diameter, and size of nano-complex. The day before the examination, the sample was prepared by drop-casting of a 5 mM aqueous solution (20 µl) dissolved in DMSO onto aluminum foils. It was coated with gold, lyophilized, and analyzed by SEM (Mira3, FEG-SEM, Tescan, USA, 5.0 kV) in a high vacuum mode.
Cell culture
A549 cell line is derived from a drug-resistant human lung carcinoma tissue and adherent in culture. This cell line was obtained from the Pasture Institute (Tehran, Iran). It was cultured in RPMI-1640 medium supplemented with 10% FBS, 100 units/ml penicillin, and 100 µg/ml streptomycin at 37 °C in 5% CO2 a humidified atmosphere. Moreover, to develop resistance, 0.1 µM Dox was added to the medium which was not toxic to the cells.
Cell viability
Cells were detached from flasks by trypsin and seeded in 96-well plates with a cell density of 1.5×104 cells/well (containing 200 μl/well of medium) and incubated for 24 hr at 37 °C in a humidified atmosphere of 5% CO2. The next day, cells were treated with different concentrations of peptides (15, 25, 50 μM), and incubated at the same condition for 24 hr. Afterward, Dox was added to plates with 20 μM concentration (IC50), and cells were incubated for the next 24, 48, 72 hr. The MTT assay was performed according to literature (37). Data were collected from at least 3 independent experiments performed in triplicates and analyzed using the Graphpad Prism 7 software package.
Cellular uptake
Fluorescent Microscopy. In order to study the cellular uptake of FAM-labeled peptides in the A549 cell line, cells were seeded in 6-well plates and incubated overnight. Then they were washed with PBS, treated with FAM-labeled peptides and nano-complexes, and incubated for 90 min at 37 °C. Next, the medium was removed and cells were washed with PBS three times. In order to fix the cells in 10 ml volume, 1 ml of formaldehyde (3.7%) was diluted with 9 ml PBS and then 2 ml of prepared fixative was added to cells. Cells were then washed with PBS three times. Samples were stored and protected from light at 4 °C until the coverslips were placed cell-side-down on cytoslides and observed using an Olympus IX81 fluorescence microscope (Olympus Optical Co., Tokyo, Japan).
Flow cytometry
First, cells were seeded in 6-well plates (3×105 cells/well) and incubated for 24 hr. Then the medium was removed and cells were washed with PBS. FAM-labeled peptides and nano-complexes were added to wells and incubation was continued for 90 min. The old medium was removed and cells were washed with PBS three times and detached by trypsin/EDTA. Then 2 ml of medium was added to each well, and cells were centrifuged for 5 min, followed by washing three times with PBS and finally suspended in flow cytometry buffer to study the cellular uptake of FAM-labeled peptides and nano-complexes.
Rhodamine123 accumulation
In order to evaluate the penetrating effect of designed peptides on rhodamine123 accumulation (as P-gp substrate), the A549 cell line was seeded in 24-well plates and incubated for 24 hr. After removing the old medium, peptides were added to wells at a concentration of 50 µM and the incubation was continued for 48 hr. Then the peptide solution was removed and cells were washed three times with PBS. Then 0.5 µM rhodamine123 solution was added to each well, and cells were incubated for 3 hr. Finally, after washing with cold PBS, cells were lysed with Triton X-100 (1%) and centrifuged at 112 g for 5 min. The fluorescence was read by a spectrometer.
ABCB1 expression assay
Real-time PCR. To simultaneously monitor the PCR reaction in real-time and amount of products (DNA, cDNA, or RNA), cells were seeded in 6-well plates and kept overnight to adhere. Then, cells were treated with 50 µM of chosen peptides (P3, P5, P7, and P8) and their nano-complexes (P3/E10, P5/E10, P7/E10, and P8/E10) and incubated for 24 hr. Four different samples were prepared for assessment consisting of peptides (P3, P5, P7, and P8), peptide-Dox conjugates (P3-Dox, P5-Dox, P7-Dox, and P8-Dox), nano-complex (P5/E10), and nano-complex-Dox (P5/E10-Dox). Cells without any treatment were considered as control. On the third day of the experiment, Dox was added to the wells, and incubation was continued for the next 48 hr. Cells were washed twice by PBS, 100 µl trypsin/EDTA was added, and incubated for 5 min. After detachment, cells were transferred to Eppendorf tubes and washed twice by PBS. After addition of 1 ml Trizol, cells were shaken by vortex and incubated for 5 min at room temperature. Then 200 µl chloroform was added to each tube and they were shaken slowly for 15 sec, placed in ice for 5 min, and centrifuged for 15 min at 16128 g, 4 °C. The supernatant was transferred to RNAase free tubes, isopropanol added, and incubated for 15 min at 4 °C. Samples were centrifuged for 15 min at 16128 g, 4 °C. After discarding the supernatant, 1 ml of cold ethanol (75%) was added to samples and they were centrifuged for 10 min at 6300g, -20 °C. 50 µl diethyl pyrocarbonate (DEPC)-treated water was added to samples. For the synthesis of the first-strand cDNA from RNA, RevertAidTM(First-strand cDNA synthesis kit, Thermo-Fisher, Waltham, MA) was employed. To this end, 1 μl 5x reaction buffer, 1 μl RiboLock, and 1 μl dNTP were added to samples. Then 1 μl Reverse Transcriptase was added to the mixture and it was kept in PCR for 60 min at 42 °C. Reaction finished by increasing the temperature to 70 °C for 10 min and the sample was transferred to ice. For approving complete synthesis of cDNA, electrophoresis was carried out. The ABCB1 expression level was also determined using the Corbett Rotor gene 6000 device and Takara Master Mix. Primers were designed and synthesized by Pishgam Company. Table 1 lists the primers and their properties. First, primers were centrifuged at 6300 g for 20 sec at 4 °C. Then DEPC-treated water was added to the diluted primers. Vials containing primers were kept at lab temperature for 30 min, centrifuged in the same condition, and kept at -20 °C.
Table 1.
The sequence of used primers and their important properties
| Primer | Sequence | T m ( ° C) | PCR product (bp) |
|---|---|---|---|
| β-actin | Forward:5CTCACCATGGATGATGATATCGC Reverse:5AGGAATCCTTCTGACCCATGC |
62.9 61.3 |
23 21 |
| ABCB1 | Forward:5GAGAGATCCTCACCAAGCGG Reverse:5ATCATTGGCGAGCCTGGTAG |
62.5 60.5 |
20 20 |
Amplification was carried out using SYBR® Premix Ex Taq™ II (RR820, USA) in a Rotor-Gene 6000 real-time PCR detection system (Corbett, UK), according to the manufacturer’s instructions. β-actin was used as a reference (housekeeping) gene to evaluate and compare the quality of different cDNA samples. Finally, before the data analysis, melting curves were assessed to prove the specific gene peak and absence of dimer primers. To achieve higher standards, the concentrations and Tm of primers were normalized. The synthesized cDNAs for all eleven samples were assessed to monitor the expression level of the ABCB1 gene.
Results
Synthesis and characterization
Eleven sequences of peptides with different patterns were manually synthesized based on the SPPS method. The ninhydrin test was implemented to monitor the synthesis process and deprotection-coupling cycle. In order to obtain the peptides for the experiment, they were cleaved from resin, lyophilized, and kept at -20 ºC. Table 2 shows all the synthetic peptides and their properties.
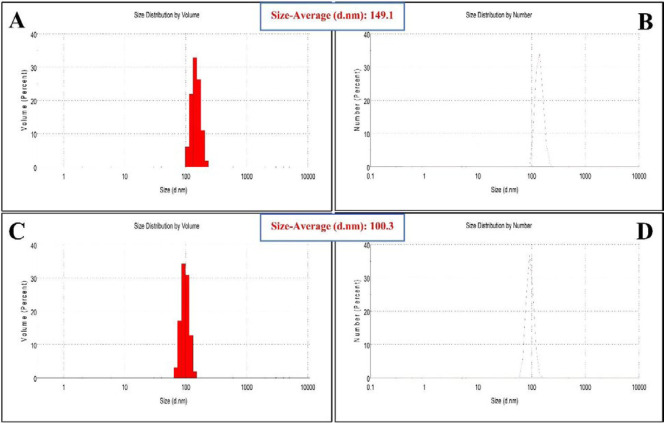
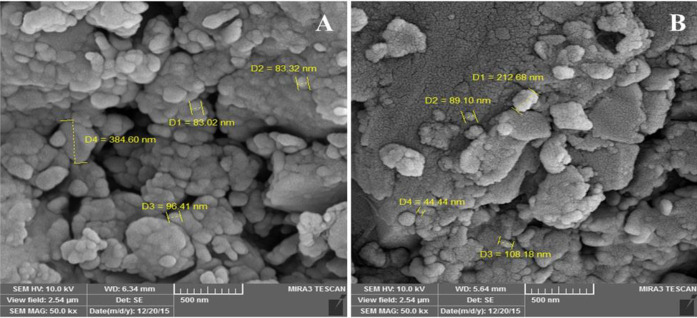
Zetasizer was used to evaluate the size distribution of obtained peptide (P3) and its nano-complex formed in the presence of E10 (P3/E10). The average size of P3 was reported to be 149.1 nm (PDI = 0.39) (Figures 1A, B), while the P3/E10 nano-complexes were smaller in size, measured to be 100.3 nm (PDI = 0.34) (Figures 1C, D). Based on SEM images, P3 showed spherical structure, while P3/E10 nano-complexes were rod-like nanostructures with smaller sizes (Figures 2A, B).
Figure 1.
Size distribution obtained by Zetasizer for P3 (A, B) and P3/E10 nano-complexes (C, D)
Figure 2.
SEM images of P3 (A) and P3/E10 nano-complexes (B)
Cell viability
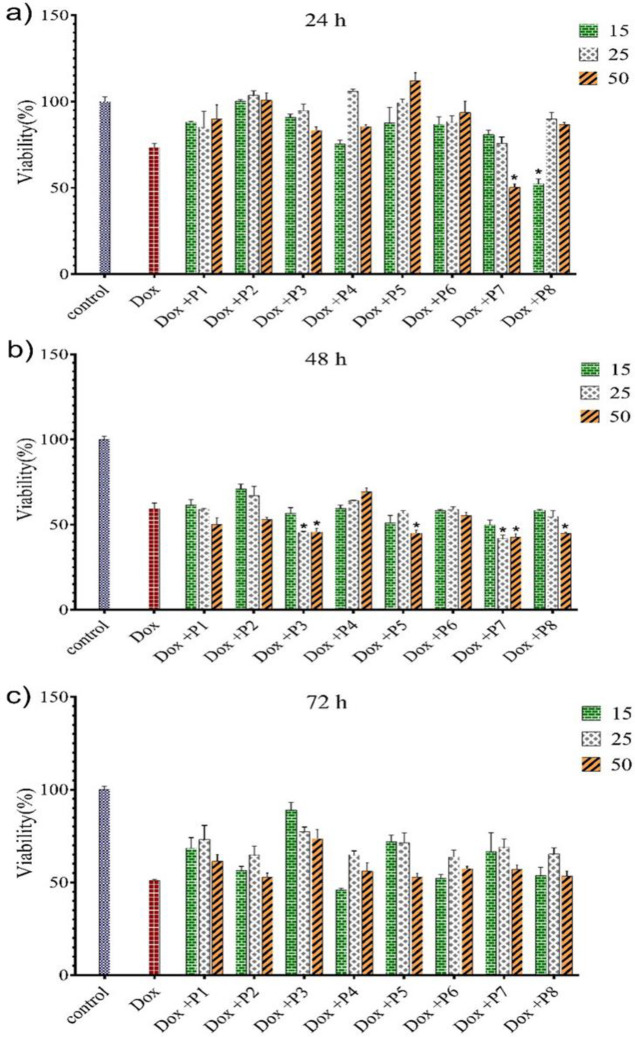
MTT assay was employed to examine the effects of CPPs on Dox cytotoxicity against A549 Dox-resistant cells at different concentrations including 15, 25, and 50 µM of CPPs. Before the experiment, IC50 of Dox was assessed, which was measured to be 20 µM. As shown in Figure 3a, at 15 µM concentration of peptides (P1-P7) and after 24 hr, there was no enhancement in toxicity of Dox in comparison with cells treated with free Dox and control, while P8 significantly reduced the cell viability to 52.5%. 25 µM and 50 µM of P7 also resulted in Dox cytotoxicity.
Figure 3.
Effects of CPPs on Dox toxicity against A549 Dox-resistant cell line after 24 hr (a), 48 hr (b), and 72 hr (c) exposure at 3 concentration levels including 15, 25, and 50 µM
CPPs: cell-penetrating peptides
After 48 hr, 50 µM of P5, P7, and P8 increased the cytotoxicity of Dox (Figure 3b). After 72 hr, compared with cells treated with free Dox and control, there was no meaningful change in the cell viability, except for the group treated with 15 µM of P4-Dox (Figure 3c).
Cellular uptake
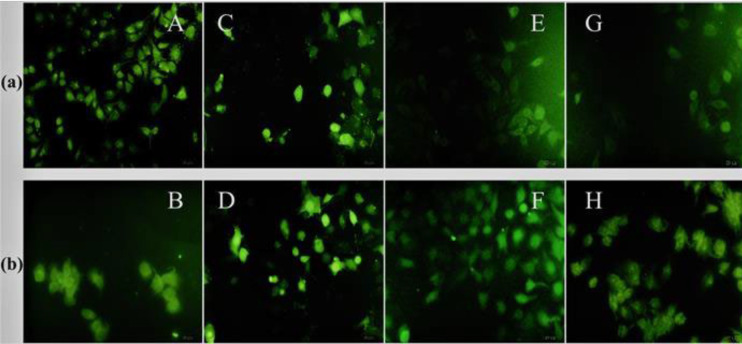
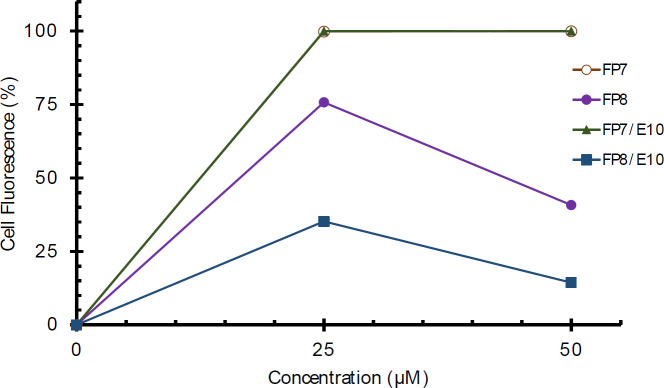
Fluorescent microscopy was utilized to examine the cellular uptake of 25 and 50 µM FAM-labeled P7 (FP7), FAM-labeled P8 (FP8), and FP7/E10 and FP8/E10 nano-complexes on A549 cells (Figures 4A, B). Based on results, at 25 µM, FR10 showed the highest uptake and its conjugation with E10 did not have any effect on its cellular uptake, whereas the nucleus uptake was slightly decreased. While the highest uptake was observed at 50 µM of FP7/E10, at both concentrations, FP8/E10 illustrated better uptake in comparison with FP8. Generally, arginine-rich peptides had better uptake than those with lysine. Furthermore, they were mostly distributed around the cell nucleus and demonstrated homogenous staining through the cell structure.
Figure 4.
Fluorescent Images of row (a): A (FP7 25 µM), C (FP7 50 µM), E (FP8 25 µM), and G (FP8 50 µM); row (b): B (FP7/E10 25 µM), D (FP7/E10 50 µM), F (FP8/E10 25 µM), and H (FP8/E10 50 µM) on A549 cells
FP7: FAM-labeled P7; FP8: FAM-labeled P8
Flow cytometry was used to quantitatively assess the cellular uptake of FR10, FK10, and their nano-complexes at the same concentrations. FP7 and FP7/E10 showed similar behavior with high efficiency in cell entrance, while FP8 and FP8/E10 showed poor cellular uptake. Furthermore, by increasing the concentration from 25 to 50 µM, the cellular uptake of FP8 was decreased (Figure 5).
Figure 5.
Cellular uptake of FP7, FP8, and FP7/E10, and FP8/E10 nano-complexes by A549 cells. The uptake was measured as the relative fraction of positive cells (%) from flowcytometry analysis of all living cells positive for the fluorophore
FP7: FAM-labeled P7; FP8: FAM-labeled P8
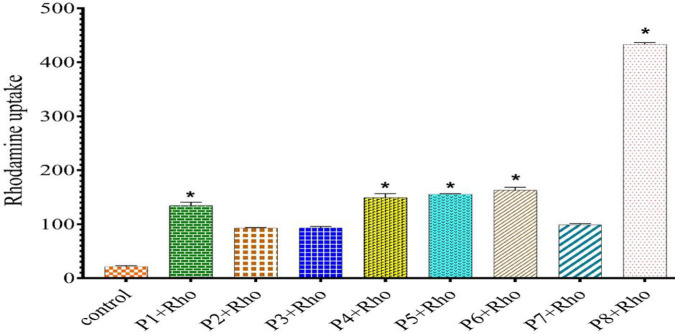
To investigate how the CPPs impact P-gp function, rhodamine123 accumulation assay was carried out (excitation and emission at 485 and 530 nm, respectively) for each sample (Figure 6). According to our results and compared with control, all CPPs could successfully increase the rhodamine123 uptake. P4, P5, and P6 had a practically similar impact on the rhodamine123 accumulation, while P8 had a significant effect on P-gp. On the other hand, compared with similar behavior of both P3 and P2, P1 led to higher accumulation. P7 was in competition with block and alternative sequences of peptides containing tryptophan and lysine.
Figure 6.
Rhodamine123 accumulation in the presence of CPPs after 48 hr incubation
ABCB1 expression assay
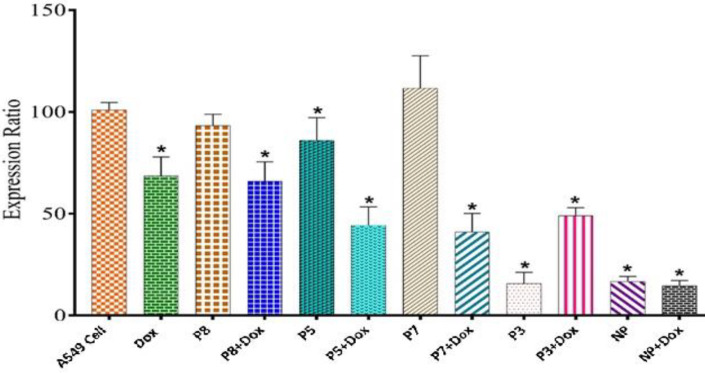
ABCB1 expression in the A549 cell line was assessed using real-time PCR. In this regard, cells were treated with the same concentrations of peptides and Dox, then the RNA was extracted and its cDNA was synthesized and normalized. Compared with controls and among all the designed CPPs, P3 had the most significant inhibitory effect on the ABCB1 gene expression, however, the Dox inhibitory behavior was not improved. Furthermore, P5-Dox and P7-Dox, significantly decreased gene expression (Figure 7). Moreover, the P5/E10 nano-complexes were employed to investigate their effects on gene expression. As a result, they either solely or in the presence of Dox, considerably reduced the expression level.
Figure 7.
ABCB1 expression level in A549 cell line after 48 hr of treatment with different peptides and peptide-Dox formulations
ABCB1: ATP-binding cassette B1
Discussion
In the present study, the morphology, size, and cellular uptake of obtained peptides were assessed by SEM, Zetasizer and fluorescence microscopy, and flow cytometry, respectively. Based on SEM images and Zetasizer analysis, the addition of E10 to peptides resulted in tangible changes to their size and morphology. SEM images of P3/E10 nano-complexes represented the creation of rod-like nanostructures that were connected to each other. The formation of these structures could be explained by the presence of electrostatic attraction (salt bridges) formed between cationic arginine/lysine residues from two different peptide chains and polyanionic polyglutamate (E10) residues (38).
There are two major mechanisms introduced for cellular uptake of CPPs including endocytosis and direct translocation (39, 40). Our results indicated higher uptake efficacy of P7 compared with P8, which might be due to the fact that the cell surface proteoglycans promote the uptake of P7 through hydrophobic interactions, while P8 experiences electrostatic interactions. In the absence of proteoglycans, arginine-rich peptides with 5 to 15 residues use other pathways to sufficiently enter the cells (41), which justifies the efficient uptake of P7. It is worth noting that, guanidinium residue is necessary for faster uptake of synthetic arginine-rich peptides (42) which is in contrast with the natural CPPs like TAT (Trans Activator of Transcription). The main mechanism for cellular uptake of CPPs is not fully understood, but studies with fluorescent microscopy and flow cytometry support the direct translocation of arginine-rich peptides into cells (35, 36, 43-49). To further study the cellular uptake of obtained peptides, fixed cells led to the nuclear distribution of peptides while in unfixed cells, they mostly distributed in cytoplasmic parts. Furthermore, we could not distinguish the amount of internalized and membrane-attached peptides by flow cytometry. In the recent study and based on the results acquired from confocal microscopy, polycationic peptides like poly K along with poly R were not taken efficiently by cells (50), while integrating poly glutamates like E8 into the structure of peptides could enhance the cellular uptake (45, 47), however, in the present study, E10 could not lead to dramatic changes in the cellular uptake of P7. Moreover, P8 had higher uptake compared with P8/E10 nano-complexes in different concentrations. On the contrary, rhodamine123 accumulation studies suggested that the lysine-rich peptides increase rhodamine123 accumulation. Dox is a hydrophobic anticancer drug with short retention time within the cells due to the robust activity of efflux pumps in cancer cells. This study represents P3, P5, P7, and P8 as promising CPPs that could increase the Dox retention time and its cytotoxic behavior in A549 Dox-resistant cells probably due to inhibition of negatively charged P-gp efflux pumps. Furthermore, CPPs can also interact with P-gp through forming hydrogen bonds and acting as its substrates (30), which extends the cellular retention time of Dox. Based on our observations, the main mechanism for cellular uptake of designed CPPs was direct translocation, which did not lead them to endosomal entrapment and degradation and probably this was the main reason for improved cytotoxic behavior of Dox at higher concentrations of CPPs. Another study used cyclic [W(WR)4]-Dox and linear [W(WR)4]-Dox (30) to increase the cellular uptake of Dox by inhibiting the efflux pumps. The cyclic structure could exhibit higher efficacy compared with the linear one (51). Based on the same mechanism and using the cationic peptide (NK-2) at 5 µM concentration, Dox resistance was reversed in drug-resistant lung cancer cell lines (52). In another study, R8-Dox complexes were prepared to evaluate their anticancer activity compared with free Dox. At 10 µM concentration and after 24 hr, R8 improved the toxicity of Dox, while their separate employment at the same concentration did not show any significant cytotoxicity. This was possibly due to the fact that R8 had high permeability and affinity for cellular DNA and RNA, resulting in Dox affinity towards targeted cells (53). In the present study, P7 also increased the Dox toxicity at concentrations of 25 and 50 µM. Furthermore, real-time PCR was applied to see the ABCB1 expression in different samples, which only shows the gene expression level and it was not clear whether the amount of the protein was increased, indicating the requirement for Western blot for complete studies. P3 could successfully decrease ABCB1 expression, while P5 unexpectedly enhanced its expression level. In the presence of Dox, the designed CPPs except P3 could decrease the gene expression level. On the other hand, P-gp expressing cells show cytoplasmic alkalization and extracellular acidification by catalyzing ATP-dependent proton extrusion (52). In fact, energetic alterations of cell membrane depolarization, low transmembrane potential, and low extracellular pH as well as perturbations in cellular ion transport are considered as characteristics of MDR cancer cells. These are favorable for cationic peptides to act against P-gp. Duan et al. evaluated the antitumor ability of paclitaxel (PTX)-conjugated TAT and LMWP (low molecular weight protamine) in comparison with free PTX on A549 cancer cells. Results indicated that PTX-LMWP has the potential for inhibiting tumor growth and considered a promising candidate to treat drug-resistant lung cancer (54). To bypass P-gp and overcome MDR cells, a hybrid CPP, comprising TAT and a drug binding motif (DBM) has been developed to deliver Dox into the K562 leukemia cell line and its P-gp overexpressing subline KD30. In this study, the endocytic cellular uptake of Dox was significantly enhanced in both cell lines, and it was supported that, this combination reduced the P-gp-mediated drug efflux (55).
Conclusion
Results showed that employment of CPPs with chemotherapeutics could improve chemotherapy efficiency via bypassing P-gp and overcoming the MDR phenomenon. Four synthesized peptides including P3, P5, P7, and P8 could successfully increase the accumulation of Dox within the drug-resistant A549 cells. PCR studies also proved the positive effects of designed peptides, in which they improved the Dox cytotoxicity by decreasing the efflux pumps gene expression. Finally, mentioned peptides are presented as effective agents to be applied with chemotherapeutics to enhance the survival rate of patients suffering from cancer.
Acknowledgment
We would like to thank the National Institute for Medical Research Development, Tehran, Iran (NIMAD, grant no. 963501) for their financial support. We appreciate the authorities of the Drug Applied Research Center and Monoclonal Antibody and Immunology Research Center of Tabriz University of Medical Sciences, Tabriz, Iran, for supporting the core facility.
Conflicts of Interest
The authors report no conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Dou L, He H, Zhang Y, Shen Q. Multifunctional nanoparticles as nanocarrier for vincristine sulfate delivery to overcome tumor multidrug resistance. Mol Pharm. 2014;11:885–894. doi: 10.1021/mp400547u. [DOI] [PubMed] [Google Scholar]
- 3.Dawar S, Singh N, Kanwar RK, Kennedy RL, Veedu RN, Zhou SF, et al. Multifunctional and multitargeted nanoparticles for drug delivery to overcome barriers of drug resistance in human cancers. Drug Discov Today. 2013;18:1292–1300. doi: 10.1016/j.drudis.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer. 2011;71:3–10. doi: 10.1016/j.lungcan.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Dou L, He H, Zhang Y, Shen Q. Multifunctional nanoparticles as nanocarrier for vincristine sulfate delivery to overcome tumor multidrug resistance. Mol Pharm. 2014;11:885–894. doi: 10.1021/mp400547u. [DOI] [PubMed] [Google Scholar]
- 6.Li W, Zhang H, Assaraf YG, Zhao K, Xu X, Xie J, et al. Overcoming ABC transporter-mediated multidrug resistance: molecular mechanisms and novel therapeutic drug strategies. Drug Resist Updat. 2016;27:14–29. doi: 10.1016/j.drup.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Niazi M, Zakeri-Milani P, Najafi Hajivar S, Soleymani Goloujeh M, Ghobakhlou N, Shahbazi Mojarrad J, et al. Nano-based strategies to overcome p-glycoprotein-mediated drug resistance. Expert Opin Drug Metab Toxicol. 2016;12:1021–1033. doi: 10.1080/17425255.2016.1196186. [DOI] [PubMed] [Google Scholar]
- 8.Dong X, Mumper RJ. Nanomedicinal strategies to treat multidrug-resistant tumors: current progress. Nanomedicine. 2010;5:597–615. doi: 10.2217/nnm.10.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yardley DA. Drug resistance and the role of combination chemotherapy in improving patient outcomes. Int J Breast Cancer. 2013;2013:15–??. doi: 10.1155/2013/137414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cukierman E, Khan DR. The benefits and challenges associated with the use of drug delivery systems in cancer therapy. Biochem Pharmacol. 2010;80:762–770. doi: 10.1016/j.bcp.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel NR, Pattni BS, Abouzeid AH, Torchilin VP. Nanopreparations to overcome multidrug resistance in cancer. Adv Drug Deliv Rev. 2013;65:1748–1762. doi: 10.1016/j.addr.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markman JL, Rekechenetskiy A, Holler E, Ljubimova JY. Nanomedicine therapeutic approaches to overcome cancer drug resistance. Adv Drug Deliv Rev. 2013;65:1866–1879. doi: 10.1016/j.addr.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livney YD, Assaraf YG. Rationally designed nanovehicles to overcome cancer chemoresistance. Adv Drug Deliv Rev. 2013;65:1716–1730. doi: 10.1016/j.addr.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Fodale V, Pierobon M, Liotta L, Petricoin E. Mechanism of cell adaptation: when and how do cancer cells develop chemoresistance? Cancer J. 2011;17:89–95. doi: 10.1097/PPO.0b013e318212dd3d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillet JP, Gottesman MM. Mechanisms of multidrug resistance in cancer. Methods Mol Biol. 2010;596:47–76. doi: 10.1007/978-1-60761-416-6_4. [DOI] [PubMed] [Google Scholar]
- 16.Mesgari Abbasi M, Valizadeh H, Hamishekar H, Mohammadnejad L, Zakeri-Milani P. The effects of cetirizine on P-glycoprotein expression and function in vitro and in situ. Adv Pharm Bull. 2016;6:111–118. doi: 10.15171/apb.2016.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callaghan R, Luk F, Bebawy M. Inhibition of the multidrug resistance P-glycoprotein: time for a change of strategy? Drug Metab Dispos. 2014;42:623–631. doi: 10.1124/dmd.113.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abd Ellah NH, Taylor L, Ayres N, Elmahdy MM, Fetih GN, Jones HN, et al. NF-[kappa]B decoy polyplexes decrease P-glycoprotein-mediated multidrug resistance in colorectal cancer cells. Cancer Gene Ther. 2016;23:149–155. doi: 10.1038/cgt.2016.17. [DOI] [PubMed] [Google Scholar]
- 19.Mohammadzadeh R, Baradaran B, Valizadeh H, Yousefi B, Zakeri-Milani P. Reduced ABCB1 expression and activity in the presence of acrylic copolymers. Adv Pharm Bull. 2014;4:219–224. doi: 10.5681/apb.2014.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanke U, May K, Rozehnal V, Nagel S, Siegmund W, Weitschies W. Commonly used nonionic surfactants interact differently with the human efflux transporters ABCB1 (p-glycoprotein) and ABCC2 (MRP2) Eur J Pharm Biopharm. 2010;76:260–268. doi: 10.1016/j.ejpb.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Sodani K, Patel A, Kathawala RJ, Chen Z-S. Multidrug resistance associated proteins in multidrug resistance. Chin J Cancer. 2012;31:58–72. doi: 10.5732/cjc.011.10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavaco MC, Pereira C, Kreutzer B, Gouveia LF, Silva-Lima B, Brito AM, et al. Evading P-glycoprotein mediated-efflux chemoresistance using solid lipid nanoparticles. Eur J Pharm Biopharm. 2017;110:76–84. doi: 10.1016/j.ejpb.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 23.Sharom FJ. ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics. 2008;9:105–127. doi: 10.2217/14622416.9.1.105. [DOI] [PubMed] [Google Scholar]
- 24.Zakeri-Milani P, Valizadeh H. Intestinal transporters: enhanced absorption through P-glycoprotein-related drug interactions. Expert Opin Drug Metab Toxicol. 2014;10:859–871. doi: 10.1517/17425255.2014.905543. [DOI] [PubMed] [Google Scholar]
- 25.Kunjachan S, Rychlik B, Storm G, Kiessling F, Lammers T. Multidrug resistance: physiological principles and nanomedical solutions. Adv Drug Deliv Rev. 2013;65:1852–1865. doi: 10.1016/j.addr.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esser L, Zhou F, Pluchino KM, Shiloach J, Ma J, Tang W-k, et al. Structures of the multidrug transporter P-glycoprotein reveal asymmetric ATP binding and the mechanism of polyspecificity. J Biol Chem. 2017;292:446–461. doi: 10.1074/jbc.M116.755884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorn CF, Oshiro C, Marsh S, Hernandez-Boussard T, McLeod H, Klein TE, et al. Doxorubicin pathways: pharmacodynamics and adverse effects. Pharmacogenet Genomics. 2011;21:440–446. doi: 10.1097/FPC.0b013e32833ffb56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren F, Shen J, Shi H, Hornicek FJ, Kan Q, Duan Z. Novel mechanisms and approaches to overcome multidrug resistance in the treatment of ovarian cancer. Biochim Biophys Acta Rev Cancer. 2016;1866:266–275. doi: 10.1016/j.bbcan.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Markman JL, Rekechenetskiy A, Holler E, Ljubimova JY. Nanomedicine therapeutic approaches to overcome cancer drug resistance. Adv Drug Deliv Rev. 2013;65:1866–1879. doi: 10.1016/j.addr.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omote H, Al-Shawi MK. Interaction of transported drugs with the lipid bilayer and P-glycoprotein through a solvation exchange mechanism. Biophys J. 2006;90:4046–4059. doi: 10.1529/biophysj.105.077743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Najafi-Hajivar S, Zakeri-Milani P, Mohammadi H, Niazi M, Soleymani-Goloujeh M, Baradaran B, et al. Overview on experimental models of interactions between nanoparticles and the immune system. Biomed Pharmacother. 2016;83:1365–1378. doi: 10.1016/j.biopha.2016.08.060. [DOI] [PubMed] [Google Scholar]
- 32.Vargas JR, Stanzl EG, Teng NN, Wender PA. Cell-penetrating, guanidinium-rich molecular transporters for overcoming efflux-mediated multidrug resistance. Mol Pharm. 2014;11:2553–2565. doi: 10.1021/mp500161z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Copolovici DM, Langel K, Eriste E, Langel Ü. Cell-penetrating peptides: design, synthesis, and applications. ACS Nano. 2014;8:1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 34.Ru Q, Shang B-y, Miao Q-f, Li L, Wu S-y, Gao R-j, et al. A cell penetrating peptide-integrated and enediyne-energized fusion protein shows potent antitumor activity. Eur J Pharm Sci. 2012;47:781–789. doi: 10.1016/j.ejps.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Farkhani SM, Johari-Ahar M, Zakeri-Milani P, Shahbazi Mojarrad J, Valizadeh H. Enhanced cellular internalization of CdTe quantum dots mediated by arginine- and tryptophan-rich cell-penetrating peptides as efficient carriers. Artif Cells Nanomed Biotechnol. 2016;44:1424–1428. doi: 10.3109/21691401.2015.1031906. [DOI] [PubMed] [Google Scholar]
- 36.Mohammadi S, Zakeri-Milani P, Golkar N, Farkhani SM, Shirani A, Shahbazi Mojarrad J, et al. Synthesis and cellular characterization of various nano-assemblies of cell penetrating peptide-epirubicin-polyglutamate conjugates for the enhancement of antitumor activity. Artif Cells Nanomed Biotechnol. 2018;46:1572–1585. doi: 10.1080/21691401.2017.1379016. [DOI] [PubMed] [Google Scholar]
- 37.Farshbaf M, Salehi R, Annabi N, Khalilov R, Akbarzadeh A, Davaran S. pH- and thermo-sensitive MTX-loaded magnetic nanocomposites: synthesis, characterization, and in vitro studies on A549 lung cancer cell and MR imaging. Drug Dev Ind Pharm. 2018;44:452–462. doi: 10.1080/03639045.2017.1397686. [DOI] [PubMed] [Google Scholar]
- 38.Meuzelaar H, Vreede J, Woutersen S. Influence of Glu/Arg, Asp/Arg, and Glu/Lys salt bridges on α-helical stability and folding kinetics. Biophys J. 2016;110:2328–2341. doi: 10.1016/j.bpj.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madani F, Lindberg S, Langel , #220 , lo , Futaki S, et al. Mechanisms of cellular uptake of cell-penetrating peptides. J Biophys. 2011;2011:414729. doi: 10.1155/2011/414729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trabulo S, Cardoso AL, Mano M, de Lima MCP. Cell-penetrating peptides—mechanisms of cellular uptake and generation of delivery systems. Pharmaceuticals. 2010;3:961–993. doi: 10.3390/ph3040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Åmand HL, Rydberg HA, Fornander LH, Lincoln P, Nordén B, Esbjörner EK. Cell surface binding and uptake of arginine- and lysine-rich penetratin peptides in absence and presence of proteoglycans. Biochim Biophys Acta Biomembr. 2012;1818:2669–2678. doi: 10.1016/j.bbamem.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Bechara C, Sagan S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013;587:1693–702. doi: 10.1016/j.febslet.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt N, Mishra A, Lai GH, Wong GCL. Arginine-rich cell-penetrating peptides. FEBS Lett. 2010;584:1806–1813. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi T, Futaki S. Current understanding of direct translocation of arginine-rich cell-penetrating peptides and Its internalization mechanisms. Chem Pharml Bull. 2016;64:1431–1437. doi: 10.1248/cpb.c16-00505. [DOI] [PubMed] [Google Scholar]
- 45.Mohammadi S, Shahbazi Mojarrad J, Zakeri-Milani P, Shirani A, Mussa Farkhani S, Samadi N, et al. Synthesis and in vitro evaluation of amphiphilic peptides and their nanostructured conjugates. Adv Pharm Bull. 2015;5:41–49. doi: 10.5681/apb.2015.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mussa Farkhani S, Asoudeh Fard A, Zakeri-Milani P, Shahbazi Mojarrad J, Valizadeh H. Enhancing antitumor activity of silver nanoparticles by modification with cell-penetrating peptides. Artif Cells Nanomed Biotechnol. 2017;45:1029–1035. doi: 10.1080/21691401.2016.1200059. [DOI] [PubMed] [Google Scholar]
- 47.Soleymani-Goloujeh M, Nokhodchi A, Niazi M, Najafi-Hajivar S, Shahbazi-Mojarrad J, Zarghami N, et al. Effects of N-terminal and C-terminal modification on cytotoxicity and cellular uptake of amphiphilic cell penetrating peptides. Artif Cells Nanomed Biotechnol. 2018;46:91–103. doi: 10.1080/21691401.2017.1414823. [DOI] [PubMed] [Google Scholar]
- 48.Zakeri-Milani P, Mussa Farkhani S, Shirani A, Mohammadi S, Shahbazi Mojarrad J, Akbari J, et al. Cellular uptake and anti-tumor activity of gemcitabine conjugated with new amphiphilic cell penetrating peptides. EXCLI J. 2017;16:650–662. doi: 10.17179/excli2017-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farkhani SM, Shirani A, Mohammadi S, Zakeri-Milani P, Shahbazi Mojarrad J, Valizadeh H. Effect of poly-glutamate on uptake efficiency and cytotoxicity of cell penetrating peptides. IET Nanobiotechnol. 2016;10:87–95. doi: 10.1049/iet-nbt.2015.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. Polyarginine enters cells more efficiently than other polycationic homopolymers. J Pept Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 51.Morris MC, Depollier J, Mery J, Heitz F, Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 52.Banković J, Andrä J, Todorović N, Podolski-Renić A, Milošević Z, Miljković Đ, et al. The elimination of P-glycoprotein over-expressing cancer cells by antimicrobial cationic peptide NK-2: The unique way of multi-drug resistance modulation. Exp Cell Res. 2013;319:1013–1027. doi: 10.1016/j.yexcr.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 53.Nakase I, Konishi Y, Ueda M, Saji H, Futaki S. Accumulation of arginine-rich cell-penetrating peptides in tumors and the potential for anticancer drug delivery in vivo. J Control Release. 2012;159:181–188. doi: 10.1016/j.jconrel.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Duan Z, Chen C, Qin J, Liu Q, Wang Q, Xu X, et al. Cell-penetrating peptide conjugates to enhance the antitumor effect of paclitaxel on drug-resistant lung cancer. Drug Deliv. 2017;24:752–64. doi: 10.1080/10717544.2017.1321060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Z, Aojula H, Clarke D. Reduction of doxorubicin resistance in P-glycoprotein overexpressing cells by hybrid cell-penetrating and drug-binding peptide. J Drug Target. 2010;18:477–487. doi: 10.3109/10611860903548347. [DOI] [PubMed] [Google Scholar]