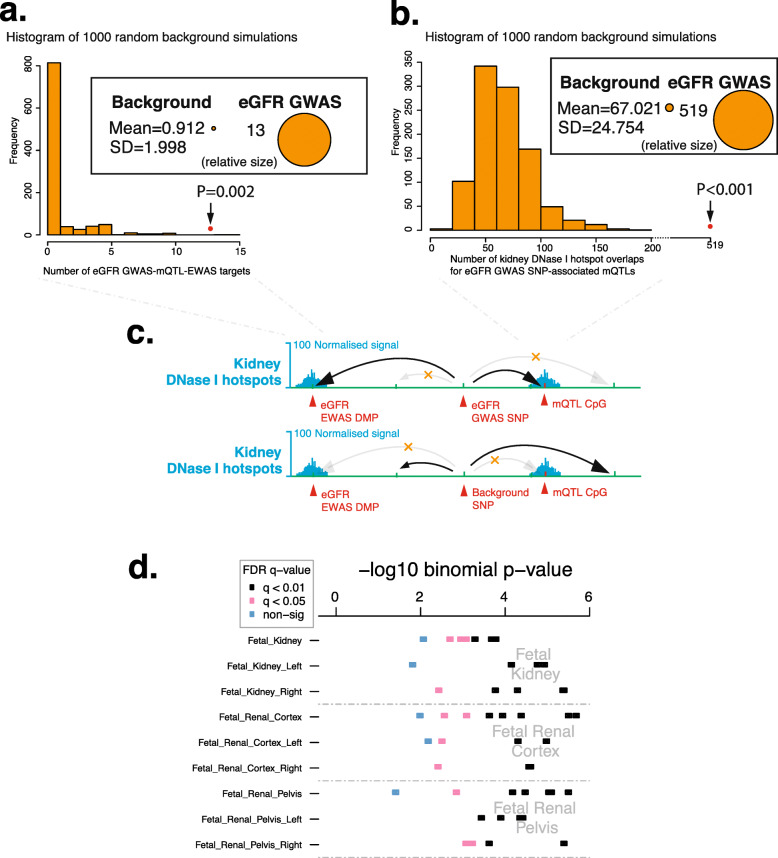
Fig. 4.
eGFR EWAS CpGs present a significant overlap with eGFR GWAS-driven meQTL effects. a Histogram of 1000 random background simulations (249 random SNPs each), for EWAS-meQTL overlap across the ARIES blood meQTL dataset (http://www.mqtldb.org/). Two hundred forty-nine unique significant SNPs from the eGFR GWAS by Hellwege et al. yield 13 SNP-meQTL-EWAS DMP sites in the Aries cohort (p = 2.0E−03, empirical test, red dot and arrow), while background SNP sets overlap a mean of 0.912 SNP-meQTL-EWAS sites. b Histogram of 1000 random background simulations (249 random SNPs each), for meQTL-kidney DNase I hotspot overlap across Roadmap Epigenomics “Kidney” sample datasets (https://egg2.wustl.edu/roadmap/web_portal/). Two thousand seven hundred thirty-three meQTL targets of 249 unique significant SNPs from the eGFR GWAS by Hellwege et al. overlap Roadmap kidney DNase I hotspots 519 times (p < 0.001, empirical test, red dot and arrow), while background SNP sets overlap Roadmap kidney DNase I hotspots a mean of 67.021 times (SD = 24.754). c Schematic showing the association of eGFR GWAS SNPs with meQTL target CpGs and eGFR EWAS CpGs (both in red text), some of which overlap kidney-specific DNase I hotspots (shown in blue, arrows indicate statistical association—not genomic contact). For comparison, a representation of a background SNP is shown. d Results from eFORGE analysis of significant ARIES meQTL CpGs associated with eGFR GWAS SNPs, indicating a higher-than expected overlap with the kidney, renal cortex, and renal pelvis DNase-seq hotspots (for additional results, see Additional file 1: Fig. S5)