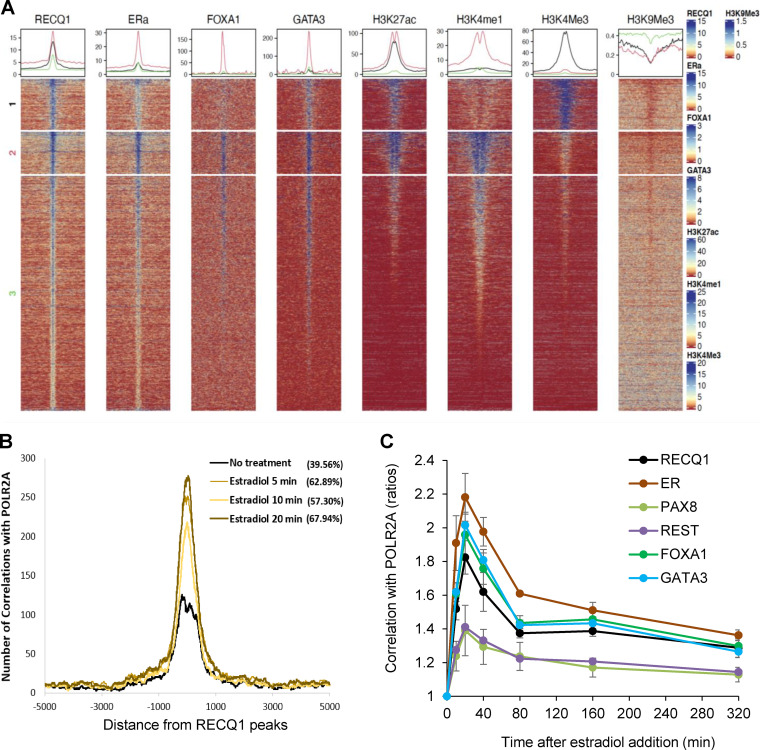
FIG 4.
Chromatin features of genome-wide RECQ1 binding. (A) Signal distribution of RECQ1, ERα, FOXA1, GATA3, H3K27ac, H3K4me1, H3K4me3, and H3K9me3 ChIP-seq data over RECQ1 peaks. The RECQ1 peaks were clustered into three groups based on H3K4me1 and H3K4me3 signal using K-means clustering. Peaks in cluster 1 correspond to strong promoters with a high H3K4me3 signal and a low H3K4me1 signal. Peaks in cluster 2 correspond to strong enhancers with a high H3K4me1 signal and a low H3K4me3 signal, with very strong RECQ1, ERα, FOXA1, and GATA3 occupancy. Cluster 3 corresponds to weak and/or inactive enhancers and promoters. (B) Colocalization of the RECQ1 binding sites (average of two replicates) with the RNA Pol II (POL2RA) binding sites. POL2RA binding sites were obtained over a time course of 0, 5, 10, and 20 min of estradiol treatment. The histogram x axis extends 5 kb upstream and 5 kb downstream from the center of the RECQ1 peaks. The extent of colocalization (percent) is measured as the fraction of RECQ1 peaks within the 5-kb window of the POLR2A peaks. (see Materials and Methods for details). (C) Graph depicting correlations between the binding sites of RECQ1, ERα, PAX8, REST, FOXA1, and GATA3 with the binding sites of POL2RA from cells collected over a time course of 0 to 320 min after estradiol treatment. Correlations were normalized using the time point t = 0. Correlations between peaks were calculated over 5-kb windows. (see Materials and Methods for details).