FIG 5.
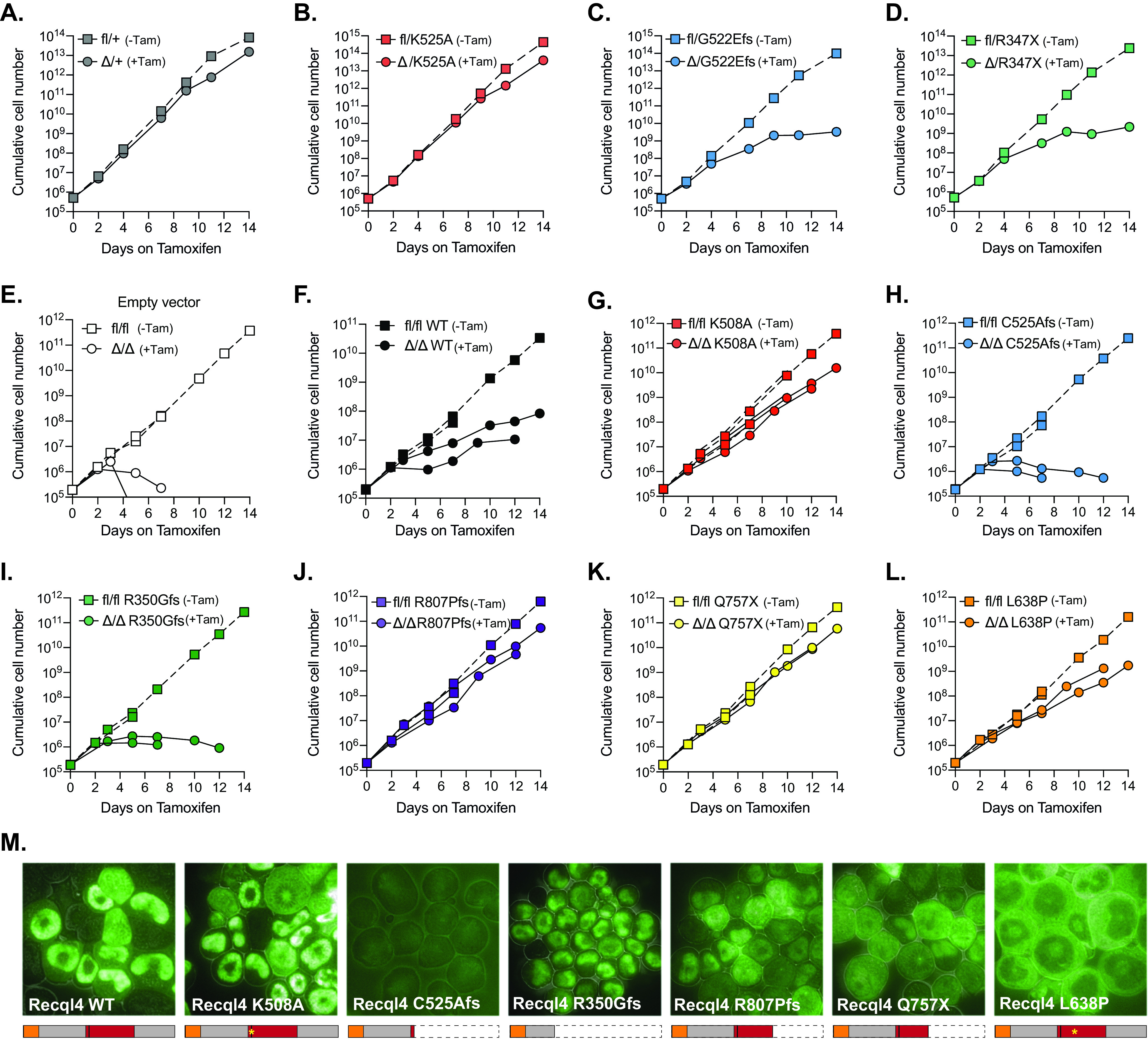
The truncating human RECQL4 mutations C525Afs and R350Gfs fail to rescue Recql4 deletion and impede proliferation, similar to their murine homologs, while mutations that conserve the protein are better tolerated. (A to D) Proliferation curves of HoxB8 immortalized R26-CreERT2 myeloid cells without (fl) and with (Δ) tamoxifen-mediated Recql4 deletion in the following cell lines: fl/+ (A), fl/K525A (B), fl/G522Efs (C), and fl/R347X (D). (E to L) Proliferation curves of HoxB8 immortalized R26-CreERT2 Recql4fl/fl control myeloid cells (E) in the presence or absence of tamoxifen and EGFP hRECQL4 overexpressing cells: wild type (F), K508A (G), C525Afs (H), R350Gfs (I), R807Pfs (J), Q757X (K), and L638P (L). Dotted lines represent individual controls not treated with tamoxifen. (M) Microscopy of EGFP-hRECQL4 expression in HoxB8 cells with WT, K508A, C525Afs, R350Gfs, R807Pfs, Q747X, and L638P. A schematic illustration of the expected protein products is outlined below each figure. The orange box represents the Sld2-like region, and the red box represents the helicase region. Cell proliferation assays using murine mutations were repeated two times using independent cell lines (replicate experiment plotted in Fig. S6). Retroviral complementation assays with human constructs were performed two times using the same parental cell line; data from each replicate are plotted separately.
