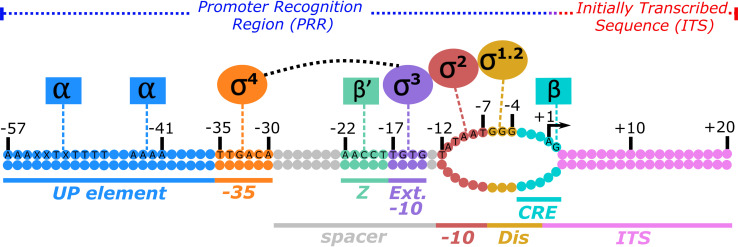
FIG 1.
Promoter sequence elements and the RNA polymerase holoenzyme subunits that recognize them. Shown is a promoter under the control of the E. coli σ70 holoenzyme. The promoter recognition region (PRR) and the initially transcribed sequence (ITS) are delineated by the transcription start site (TSS) at nucleotide position +1 (arrow). While it varies depending upon NTP availability and promoter context, a bias for purines (A or G) is typically observed for the TSS (157, 228–231). An open DNA bubble is formed around the TSS, corresponding to positions −11 to +2. Specific sequences of the nontemplate strand are listed and correspond to either the optimal and/or consensus sequence for a given element. PRRs include (i) the full upstream (UP) element (A−57AAXXTXTTTTnnAAAA−41) (101), where X is an A or T nucleotide and n is no preference, contacting the carboxyl-terminal domain of the α-subunits (αCTDs, blue), (ii) the consensus −35 hexamer (T−35TGACA−30) contacting σ4 (orange), (iii) the spacer region between −35 and −10 sites with an optimal length of 17 bp (gray) containing both (iv) the zipper (Z) element (A−22ACCT−18) (170) contacting β′ (light green) and (v) an optimal extended (Ext) −10 element (T−17GTG−14) contacting σ3 (purple), (vi) the consensus −10 hexamer (T−12ATAAT−7) contacting σ2, where the −12 bp remains double stranded (red), (vii) an optimal discriminator (Dis) element (G−6GG–4) contacting σ1.2 (yellow), and (viii) the core recognition element (CRE) (54) contacting β (teal). The CRE also makes up part of the ITS (pink), extending to the +2G position.