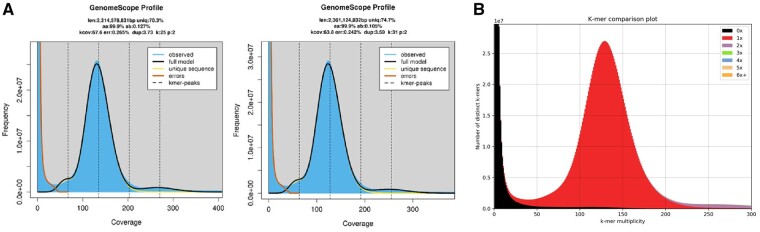
Figure 2.
(A) GenomeScope2 k-mer (25 and 31) distribution displaying estimation of genome size (len), homozygosity (aa), heterozygosity (ab), mean k-mer coverage for heterozygous bases (kcov), read error rate (err), the average rate of read duplications (dup), k-mer size used on the run (k:), and ploidy (p:). (B) Margaritifera margaritifera genome assembly assessment using KAT comp tool to compare the Illumina PE k-mer content within the genome assembly. Different colours represent the read k-mer frequency in the assembly.