Porphyromonas gingivalis is a keystone pathogen contributing to periodontitis in humans, leading to tooth loss. The oral microbiota is essential in this pathogenic process and changes from predominantly Gram-positive (health) to predominantly Gram-negative (disease) species. P. gingivalis uses its type IX secretion system (T9SS) to secrete and conjugate virulence proteins to anionic lipopolysaccharide (A-LPS).
KEYWORDS: Porphyromonas gingivalis, lipopolysaccharide exchange, virulence, periodontitis, type IX secretion system
ABSTRACT
Porphyromonas gingivalis, a bacterial pathogen contributing to human periodontitis, exports and anchors cargo proteins to its surface, enabling the production of black pigmentation using a type IX secretion system (T9SS) and conjugation to anionic lipopolysaccharide (A-LPS). To determine whether T9SS components need to be assembled in situ for correct secretion and A-LPS modification of cargo proteins, combinations of nonpigmented mutants lacking A-LPS or a T9SS component were mixed to investigate in trans complementation. Reacquisition of pigmentation occurred only between an A-LPS mutant and a T9SS mutant, which coincided with A-LPS modification of cargo proteins detected by Western blotting and coimmunoprecipitation/quantitative mass spectrometry. Complementation also occurred using an A-LPS mutant mixed with outer membrane vesicles (OMVs) or purified A-LPS. Fluorescence experiments demonstrated that OMVs can fuse with and transfer lipid to P. gingivalis, leading to the conclusion that complementation of T9SS function occurred through A-LPS transfer between cells. None of the two-strain crosses involving only the five T9SS OM component mutants produced black pigmentation, implying that the OM proteins cannot be transferred in a manner that restores function and surface pigmentation, and hence, a more ordered temporal in situ assembly of T9SS components may be required. Our results show that LPS can be transferred between cells or between cells and OMVs to complement deficiencies in LPS biosynthesis and hemin-related pigmentation to reveal a potentially new mechanism by which the oral microbial community is modulated to produce clinical consequences in the human host.
IMPORTANCE Porphyromonas gingivalis is a keystone pathogen contributing to periodontitis in humans, leading to tooth loss. The oral microbiota is essential in this pathogenic process and changes from predominantly Gram-positive (health) to predominantly Gram-negative (disease) species. P. gingivalis uses its type IX secretion system (T9SS) to secrete and conjugate virulence proteins to anionic lipopolysaccharide (A-LPS). This study investigated whether components of this secretion system could be complemented and found that it was possible for A-LPS biosynthetic mutants to be complemented in trans both by strains that had the A-LPS on the cell surface and by exogenous sources of A-LPS. This is the first known example of LPS exchange in a human bacterial pathogen which causes disease through complex microbiota-host interactions.
INTRODUCTION
Porphyromonas gingivalis is a Gram-negative pathogen linked with the progression of human periodontitis. This is a progressive inflammatory disease of the tooth’s supporting tissues in which the bone and other tissues are destroyed, ultimately leading to the loss of teeth. P. gingivalis is responsible for transforming the periodontal microbiota into a dysbiotic community and directs destructive inflammation early in the disease process at low numbers, and it is therefore considered a keystone species in the development of periodontitis (1–3). P. gingivalis appears in greater numbers with restricted species diversity of predominantly Gram-negative species later in biofilm development, when the polymicrobial infection is well established (4).
Major virulence factors of P. gingivalis contributing to this disease are the cysteine proteases known as gingipains (5, 6). These, together with approximately 30 other proteins, are secreted to the cell surface and anchored to the outer membrane (OM) by transpeptidation, where their conserved C-terminal domain (CTD) is cleaved and the new C terminus covalently attached to anionic lipopolysaccharide (A-LPS) (7, 8). This secretion and anchorage of proteins is performed by the type IX secretion system (T9SS), which has been found only in members of the Bacteroidetes/Chlorobi phyla (9–13). Substrates of the T9SS require an N-terminal signal peptide for secretion across the inner membrane (IM) by the Sec translocon and require the CTD for secretion across the OM and anchorage by the T9SS (14), and hence, substrates have been referred to as CTD proteins, cargo proteins or T9SS substrates. Cargo proteins are also greatly enriched in the outer membrane vesicles (OMVs) of P. gingivalis (15, 16).
Many of the genes required for secretion and anchorage by the T9SS have been identified and fall into three broad groups. The first group include genes/gene products involved in the biosynthesis of A-LPS. The polysaccharide portion of A-LPS is an anionic polysaccharide proposed to consist of a phosphorylated branched mannan (17, 18); however, accumulating evidence suggests that A-LPS includes a more conventional polysaccharide requiring six specific glycosyl transferases for its assembly (WbaP, GtfC, GtfE, GtfB, VimF, and GtfF) (19–24). In addition, the novel A-LPS linking sugar, which is bonded to the cargo proteins, was recently identified to be an N-seryl-N-acetylglucuronamide that is synthesized by the Wbp/Vim pathway, comprising WbpA, UgdA, WbpB, WbpE (also known as PorR), WbpD, VimE, and VimA (25–28). Genes required for both conventional O-LPS and A-LPS include those involved in core synthesis (rfa and gtfG) (18, 29, 30) and O-antigen assembly onto the lipid A core (wzx, wzz, wzy, and waaL) (20). The second group of genes are involved in signal transduction and gene regulation (porY, porX, and sigP) (9, 31, 32).
The third group are genes that code for components of the T9SS machinery, including IM proteins (PorL and PorM) and OM proteins (PorT, PorK, PorN, Sov, PorW, PorP, PorV, PorU, PorQ, PorZ, PorF/PG0534, PorE/PG1058, and PorG/PG0189) (7, 9, 33–42). While most of these components are involved in the translocation step, PorV, PorU, PorQ, and PorZ form the OM attachment complex (38), which is required for CTD cleavage and covalent attachment of A-LPS to secreted cargo proteins (7, 8, 38, 43). PorU is the CTD signal peptidase/sortase that cleaves the CTD and covalently links A-LPS to the new C terminus by transpeptidation (7, 8). The structure of PorZ contains two β-propeller domains (43) and is proposed to bind A-LPS (38). PorU and PorZ are special T9SS cargo proteins having variant CTD sequences which are uncleaved and are not modified by A-LPS but are still detected on the cell surface (7, 43). PorU and PorZ are anchored to the OM through binding to the OM β-barrel proteins PorV and PorQ, respectively, and can form separate subcomplexes in certain mutants where PorU directly binds PorV and PorZ directly binds PorQ (16, 38). In the porU mutant, PorV was observed to bind many unmodified cargo proteins in separate PorV-cargo protein complexes. These complexes were detected on the cell surface, and it was proposed that PorV acts as a cargo protein shuttle between the T9SS pore and the attachment complex for A-LPS modification (38). By cryo-electron microscopy, SprA, the Sov homologue in Flavobacterium johnsoniae, was found to form a large OM β-barrel and is proposed to be the T9SS pore (39). Sov functions by interchangeably interacting with PorV at its lateral opening and the Plug protein at its periplasmic opening. This led to the proposition that Sov transports cargo proteins to PorV through its OM channel by an alternating access mechanism.
Mutants for all three groups of genes lack black pigmentation on blood agar due to the absence of cell surface-attached Kgp and RgpA gingipains, rendering them unable to extract and produce the black pigment, μ-oxo bisheme, from hemoglobin (44). In addition, the A-LPS mutants are less able to bind the pigment to the cell surface due to the absence of A-LPS (30). Group 1 mutants that are unable to synthesize A-LPS are unable to modify their cargo proteins with A-LPS and attach them to the cell surface; instead, the cargo proteins are released into the culture milieu (26, 27). Analysis of the C-terminal cleavage of released cargo proteins in mutants lacking wbaP showed that in the absence of A-LPS, the CTD is still cleaved by the PorU sortase, suggesting that the T9SS is still responsible for their secretion (8). In contrast, cargo proteins of T9SS component mutants (group 3), although also not modified with A-LPS, vary in the extent to which they reach the cell surface. Mutants with mutations in sov (T9SS pore) porL, porM, porK, porN, porV, and porQ do not show evidence of cargo proteins on the cell surface; however, cargo proteins still reach the cell surface in porU and porZ mutants (7, 9, 38).
Many bacterial components, such as DNA, OM and periplasmic proteins, LPS and lipoproteins, and small signaling molecules, can be exchanged between strains and/or species by OMV delivery (45–50). OMVs from Gram-negative Shigella and Pseudomonas species can tightly bind/fuse with cells from other Gram-negative species, such as Salmonella species and Escherichia coli. LPS has been shown to be exchanged between Myxococcus xanthus cells indirectly by complementation of social motility and sporulation defects due to an LPS biosynthesis mutation (50).
To better understand how the OM components of the T9SS machinery function, this study investigated whether they could be complemented in trans. We found that OM proteins of the T9SS were unable to be exchanged for mutation complementation; however, A-LPS from the donor T9SS mutant (or OMVs) could be transferred to the OM of the A-LPS biosynthetic mutant cell for covalent ligation to T9SS cargo proteins.
RESULTS
Complementation of black pigmentation occurs only between an A-LPS mutant and a T9SS mutant.
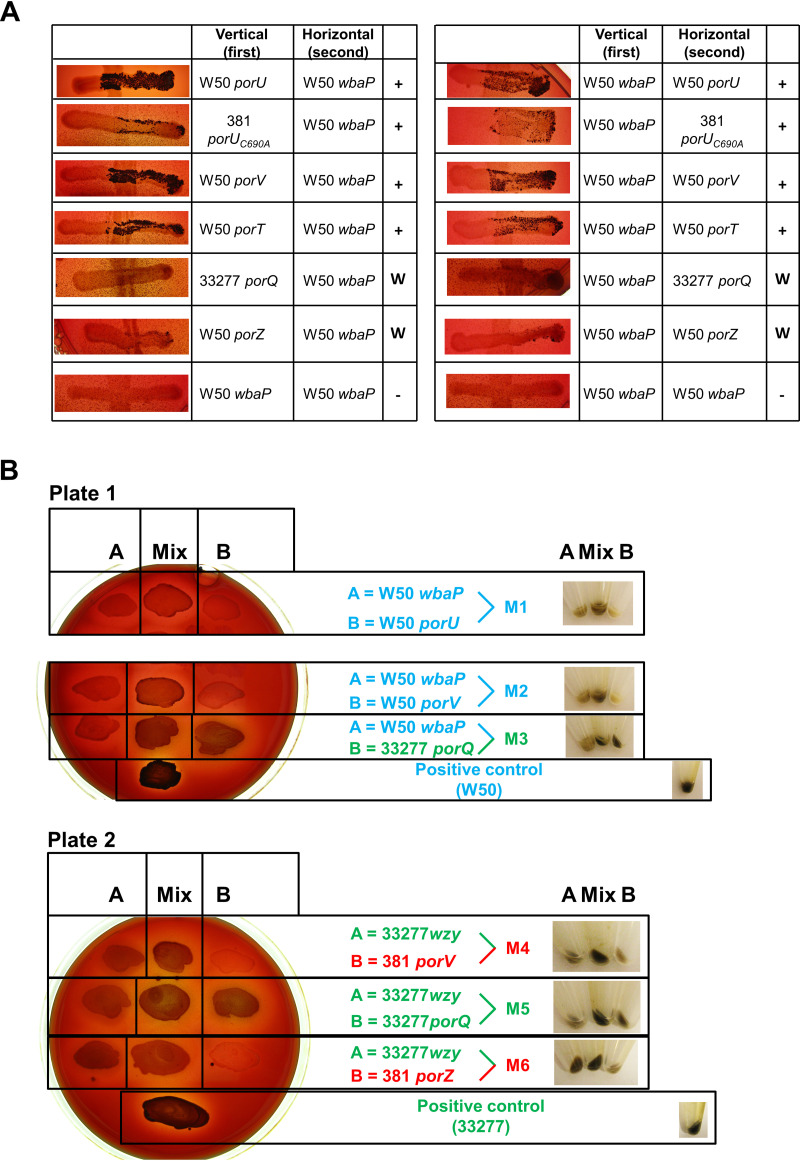
To investigate which mutants were able to complement and restore T9SS function, all possible combinations of two mutants were tested by growth on blood agar. Of all combinations, only those including a wbaP strain with a T9SS mutant could produce the black pigmentation in the right arm of the cross where both strains were mixed (Fig. 1A). In contrast, all combinations of only T9SS mutants (porT, porV, porU, porUC690A, porZ, and porQ mutants) did not produce black pigmentation (see Fig. S1 in the supplemental material). This plate pigment complementation experiment was repeated, producing similar results (data not shown). This black pigment complementation was confirmed for a selection of T9SS mutants with either wbaP or wzy by patching the two strains on BHI-T blood agar (see Materials and Methods) as single and mixed strains. Then, after 6 days’ growth, the cells from these patches were spun down, showing that the pellets from the mixed strains were darker than those from each single strain (Fig. 1B).
FIG 1.
Pigmentation complementation of mixed strains on blood agar. (A) Reciprocal crossovers of two strains between W50 wbaP and T9SS mutants. The first strain was applied vertically and the second strain was applied in one horizontal motion from left to right on BHI-T BA plates, and plates were incubated for 14 days anaerobically. +, obvious pigmentation; W, weak pigmentation; −, no pigmentation. (B) Two single strains (A and B) were patched on BHI-T BA, and a mix of the two strains was patched in the middle. Three pairs of strains were tested on each plate. The wbaP strain was used as strain A for plate 1, and the wzy strain was used as strain A for plate 2. Strain B was variable for both plates, as indicated. After 6 days’ growth, cells in patches were scraped into 1 ml of PBS and centrifuged to pellet cells. Pellets in centrifuge tubes are shown on the right. All strain names are colored according to strain background to aid matching with Fig. 2.
The size distribution of A-LPS ladders is strain specific.
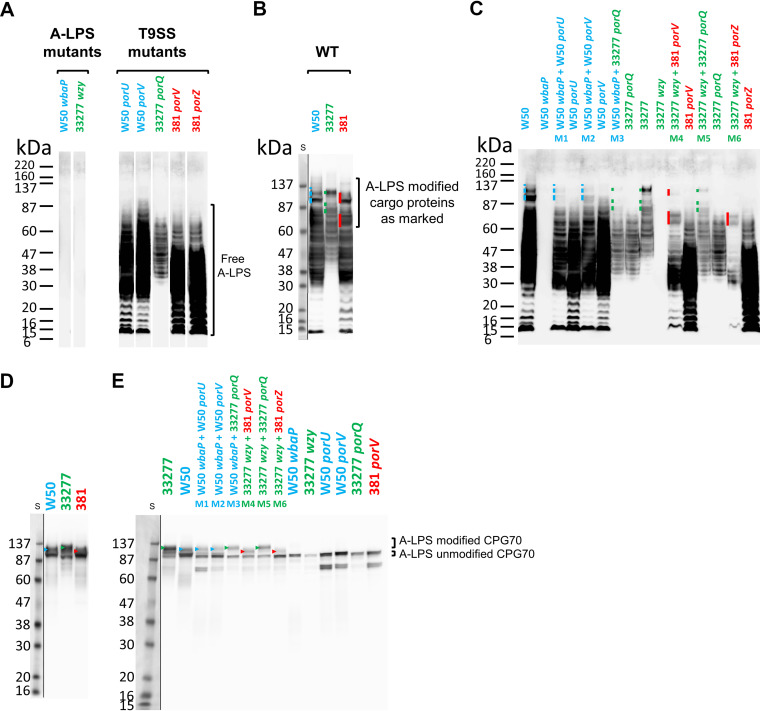
To examine if the black pigmentation was due to covalent linkage of A-LPS to T9SS cargo proteins, first, the size distribution of A-LPS within the various strains used was analyzed by Western blotting analysis using the anti-A-LPS monoclonal antibody (MAb) 1B5, which detects both free and protein-modified A-LPS (7, 37, 38). The wbaP and wzy mutants were both negative for A-LPS as expected, while the mutants lacking components of the T9SS produced free A-LPS over a broad range of molecular weights (MW) (Fig. 2A). The W50 mutants, with mutations in porU and porV, exhibited dark staining from ∼25 to 70 kDa, and the 33277 porQ mutant was more lightly stained and produced a ladder spanning from ∼35 to 65 kDa, while 381 porV and 381 porZ showed the darkest staining between ∼15 and 50 kDa (Fig. 2A).
FIG 2.
Complementation of pigmentation on blood agar correlates with covalent linkage of protein to A-LPS. Whole-cell lysates of strains were grown for 6 days on plates as either single strains or mixes of an A-LPS mutant (W50 wbaP or 33277 wzy) with a T9SS mutant (W50 porU, W50 porV, 33277 porQ, 381 porV, or 381 porZ) as shown in Fig. 1B except for the WT strains (W50, 33277, and 381, shown in separate panels), which were similarly grown and processed. All strain names are colored according to strain background. (A, B, and C) Anti-A-LPS Western blot analyses. (D and E) Anti-CPG70 Western blot analyses. Profiles of high-MW A-LPS modified proteins are indicated by a colored line or arrowhead to the left of the band(s) and color coded according to the background strain (blue, W50; green, 33277; red, 381). S, protein size ladder. The resulting image of each of the Western blots showing the protein standard is produced by two exposures of the same blot perfectly overlapped and cropped to reveal the prestained protein ladder.
In the wild-type (WT) strains, most of the A-LPS is conjugated to cargo proteins, which can be seen by the appearance of new bands of higher MW (Fig. 2B, new bands indicated by color-coded lines). When the WT strains were probed with anti-CPG70, which detects both unmodified and A-LPS-modified forms of CPG70, the strain-specific size of the A-LPS modification on the CPG70 cargo protein was evident, with strain 33277 producing the highest-MW band, followed by W50 and then 381 (Fig. 2D). Note that the size of CPG70 calculated from the amino acid sequences is the same in each strain.
Complementation between mutant strains involves covalent linkage of A-LPS to T9SS cargo proteins.
Having characterized the profiles of A-LPS in each strain for both free A-LPS and protein-conjugated A-LPS, we examined the cell pellets of the “mixed” strains (Fig. 1B) to determine whether complementation involved the covalent linkage of A-LPS to T9SS cargo proteins. The Western blots of each mixed strain sample were run in a lane near the relevant WT and T9SS mutant strains so that the appearance of new bands corresponding to A-LPS modified proteins could be closely examined (Fig. 2C). It was observed that the high-MW A-LPS forms (marked with colored bars) were only observed for the “mixed” mutant pair lysates (M1 to M6) and WT strains, demonstrating that covalent linkage of A-LPS to cargo proteins had occurred in the mixed mutant cultures (Fig. 2C). Although the new high-MW bands were sometimes weakly stained, the overall staining of the mixed strain samples was weaker than that of the corresponding T9SS mutant, demonstrating that the detection of the new bands was not due to increased sample loading. To further confirm this, one specific cargo protein was analyzed using the same samples and probed with anti-CPG70 (Fig. 2E). The mixed strain samples all produced high-MW forms that were absent in the single mutants, confirming that A-LPS modification of CPG70 had taken place. The upper A-LPS-modified CPG70 bands were of various sizes and consistent with the strain-specific results of the WT strains, where 381 had the smallest and 33277 the largest A-LPS modification (Fig. 2D).
The high-MW A-LPS patterns (Fig. 2B and C) and the A-LPS-modified CPG70 (Fig. 2D and E) appeared to follow the same pattern as the corresponding WT parent strain of the putative A-LPS donor mutant strain rather than that of the WT parent strain pattern of the wbaP or wzy mutants. Mixes M1 (A-LPS donor, W50 porU) and M2 (donor, W50 porV) corresponded to a WT W50 anti-A-LPS profile, M3 and M5 (donor, 33277 porQ) corresponded to a 33277 WT anti-A-LPS profile, and M4 (donor, 381 porV) and M6 (donor, 381 porZ) corresponded to a WT 381 anti-A-LPS profile, although the top two bands for the 381 porZ donor mix were not detected, perhaps due to a weaker complementation that was also observed on blood agar plates in Fig. 1A. The same trend is also observed for modified CPG70 for these mixes (see color-coded arrowheads in Fig. 2D and E) where the size difference of modified CPG70 corresponded to the size of the donor strain A-LPS. These results are consistent with transfer of free unconjugated A-LPS from the donor strain to the A-LPS biosynthetic mutant (wbaP or wzy), which uses the free A-LPS to conjugate to its cargo proteins. This finding rules out the alternative hypothesis that the wbaP or wzy genes or their gene products were donated to restore A-LPS biosynthesis within the recipient strain.
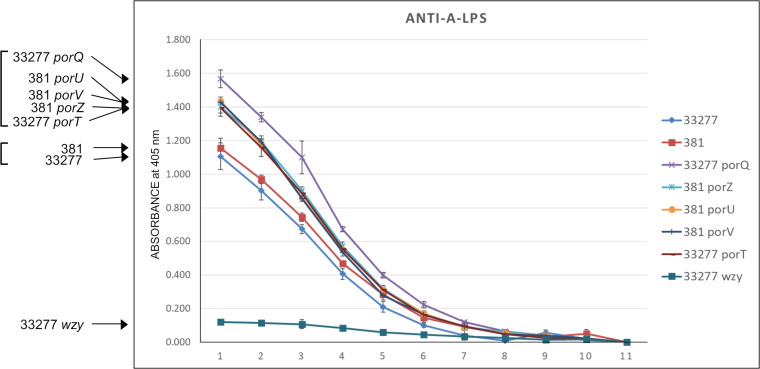
Cell surface forms of A-LPS were assessed by whole-cell enzyme-linked immunosorbent assay (ELISA) (Fig. 3). This confirmed that the wzy mutant lacked surface A-LPS. Conversely, unconjugated A-LPS was present on the surface of all tested T9SS mutants (porQ, porU, porV, porZ, and porT). These mutants produced slightly higher signals than the WT strains, 33277 and 381, which possessed predominantly protein-conjugated forms of A-LPS.
FIG 3.
Anti-A-LPS whole-cell ELISA. Anti-A-LPS dilution 1, corresponding to horizontal axis point 1, represents 1/100 dilution, followed by serial 2-fold dilutions for points 2 to 10. Point 11 represents the controls (secondary anti-mouse IgG horseradish peroxidase conjugate only) which all values were normalized to for each strain. Signal intensity order for strains is indicated on the left, and apparent clustering is indicated by brackets.
Exogenous sources of A-LPS can complement A-LPS biosynthetic mutants.
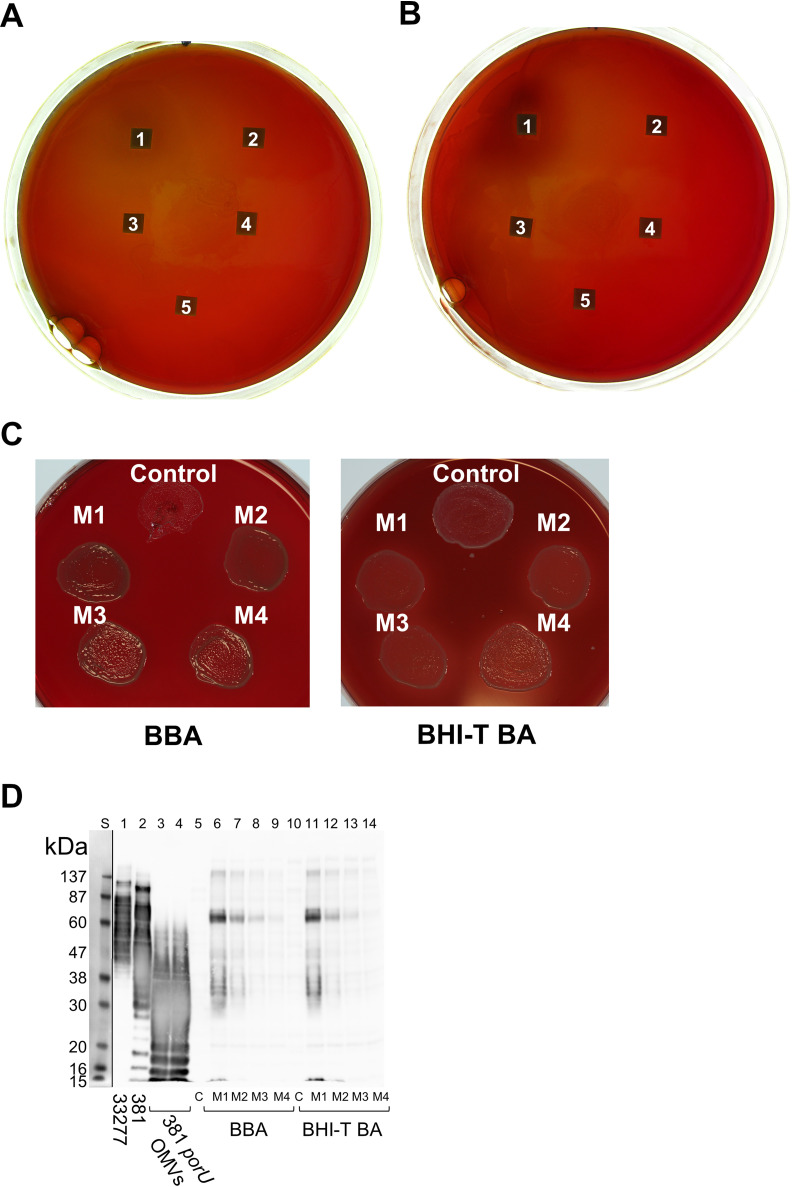
To test if cell-free forms of A-LPS could complement the defect in the wzy mutant, purified LPS from P. gingivalis containing unconjugated A-LPS was applied to sterile blotting paper on blood agar medium seeded with lawns of wzy mutant cells at two different dilutions in BHI-T growth medium. After growth, weak pigmentation could be observed around the blotting paper containing the largest amount of purified A-LPS (Fig. 4A and B).
FIG 4.
Complementation of pigmentation occurs using exogenous sources of A-LPS. (A and B) Complementation of pigmentation with ultrapurified P. gingivalis LPS. Two different dilutions of 33277 wzy, 1:4 (A) and 1:9 (B), were spread onto BHI-T BA plates. Square pieces of blotting paper were then placed on top, and 20-μl volumes of ultrapurified LPS from P. gingivalis were applied (1, 20 μg; 2, 2 μg; 3, 0.2 μg; 4, 20 ng; 5, 2 ng). Plates were anaerobically incubated for 15 days at 37°C. Pigmentation is observed surrounding blotting paper 1 on both plates. (C and D) Complementation of an A-LPS biosynthetic mutant using purified OMVs from 381 porU. (C) The control patch was 10 μl of strain 33277 wzy mixed with 10 μl of control sterile water and spread onto either BBA or BHI-T BA plates. M1, M2, M3, and M4 are from mixes (10 μl-10 μl) of the wzy strain with serial 2-fold dilutions of purified filtered OMVs from the 381 porU mutant diluted in sterile water starting at 200 μg/μl. Control and mixes were applied to plates and incubated anaerobically for 4 days. (D) Whole-cell lysates were subjected to SDS-PAGE, transferred to nitrocellulose, and immunostained with anti-A-LPS. Lane 1, WT 33277; lane 2, WT 381. Lanes 3 and 4 are equivalent to starting amounts of 381 porU OMVs; lanes 5 to 14 are whole-cell lysates from cells scraped from growth media shown in panel C. Because cell pellets were resuspended to the same wet weight per milliliter, lanes 3 and 4 represent the minimum and maximum starting amounts of porU OMVs in the mixed lysates (M1, M2, M3, and M4). The resulting image of the Western blot is produced by two exposures of the same blot perfectly overlapped and cropped to reveal the prestained protein ladder.
Another exogenous source of unconjugated A-LPS was purified filtered OMVs from the 381 porU mutant that was mixed with the 33277 wzy mutant, patched onto BBA (see Materials and Methods), which did not contain a trypsin-cleaved source of proteins, or onto BHI-T BA, which contained trypsin-cleaved soy protein fragments, and grown for 4 days (Fig. 4C). Weak pigmentation was observed for all mixes with OMVs but not for the controls without OMV material. Interestingly, on the BBA plate, OMVs were required to assist the growth of the wzy mutant. Cells from patches grown for 4 days were scraped off plates, lysed, and subjected to anti-A-LPS Western blot analysis (Fig. 4D). The control wzy lysates, as expected, showed a complete absence of A-LPS, but the lysates from cells mixed with OMVs from the porU mutant contained predominantly high-MW A-LPS modified proteins of a restricted size. There was little detectable free A-LPS; therefore, it appeared that the finite amount of donated A-LPS added was almost completely converted to a protein-conjugated form. It was also different from what was produced when cells rather than OMVs were used for the complementation, where, in general, both high-MW A-LPS-conjugated proteins and low-MW free A-LPS were observed (Fig. 2B). Due to the limited amount of A-LPS added for this complementation, it was expected that only the most abundant CTD proteins would be observed.
Coimmunoprecipitation of T9SS cargo proteins with anti-A-LPS and MaxQuant iBAQ quantitation.
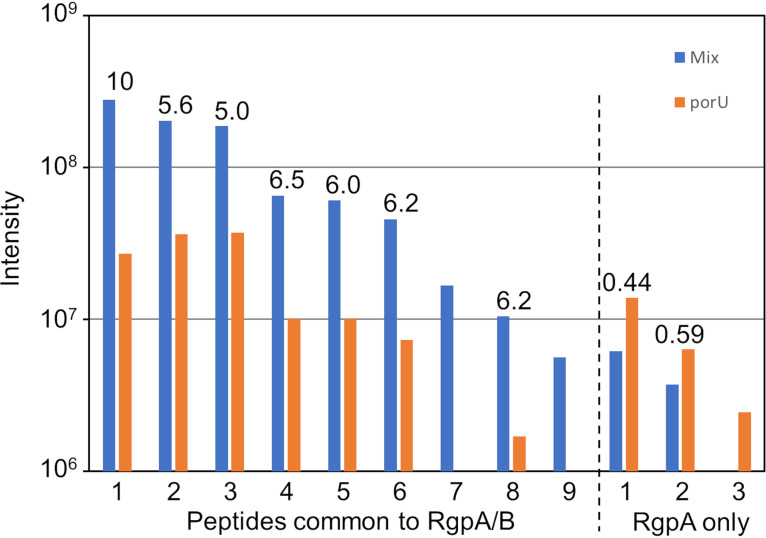
To further support that the mechanism of complementation involves the A-LPS modification of T9SS cargo proteins, the cell lysate obtained after 33277 wzy was grown with 381 porU was subjected to coimmunoprecipitation (co-IP) using anti-A-LPS. For negative controls, cell lysates from the individual strains were used. The three co-IP samples were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and quantitated by MaxQuant software using the 33277 database. Comparison of the label-free quantitation (LFQ) intensity of the proteins in the mixed sample with the maximum LFQ intensity in the individual strains revealed 10 highly enriched proteins (Table 1). Six of these proteins are T9SS cargo proteins, including HBP35, which was enriched by more than 100-fold. Quantitation of the amount of protein pulled down using iBAQ (intensity-based absolute quantification) revealed that HBP35 and RgpA/RgpB were easily the most abundant (Table 1). The strong preference for pulling down T9SS cargo proteins in the mixed sample confirms that A-LPS modification of cargo proteins occurs during complementation. To determine which form of Rgp was pulled down, the individually quantitated peptides were examined. Seven of the nine peptides common to RgpA and RgpB were quantifiable. The other two peptides were not, as they were not detected in the control sample (Fig. 5). The intensity ratios for these quantifiable peptides were very similar, varying from 5.0 to 10.3, indicating a significant enrichment in the mixed sample, whereas the 3 peptides specific to RgpA exhibited ratios of <0.6. The clear demarcation between these two groups suggests that only RgpB was enriched. The MaxQuant analysis was repeated without RgpA in the database so that the enrichment and abundance of RgpB could be better estimated. The “inferred” RgpB was enriched by 6-fold and was the most abundant protein that was specifically pulled down (Table 1). The values for RgpB are likely to be underestimates, since the protein detected in the negative controls is inferred to be mostly RgpA rather than RgpB, since there were similar abundances for both RgpA-specific and RgpA/RgpB-common peptides.
TABLE 1.
MaxQuant iBAQ quantitation of proteins coimmunoprecipitated with anti-A-LPS from a mix of 33277 wzy and 381 porU strainsa
| Protein name | Locus tagd | LFQ intensity |
LFQ ratio | iBAQ abundance | No. of peptides (mix) | |
|---|---|---|---|---|---|---|
| Mix | Negative | |||||
| RgpBb | PGN_1466 | 7.6E+08 | 1.27E+08 | 6.0 | 2.5E+07 | 9 |
| HBP35 | PGN_0659 | 2.5E+08 | 2.4E+06 | 106.4 | 1.4E+07 | 7 |
| RgpAc | PGN_1970 | 7.8E+08 | 1.4E+08 | 5.4 | 1.2E+07 | 11 |
| RgpB | PGN_1466 | 7.8E+08 | 1.4E+08 | 5.4 | 1.2E+07 | |
| PPAD | PGN_0898 | 8.0E+07 | 0 | 2.7E+06 | 10 | |
| Unknown function | PGN_1321 | 1.3E+07 | 0 | 6.3E+05 | 3 | |
| Unknown function | PGN_1366 | 2.5E+07 | 9.6E+05 | 26.3 | 6.2E+05 | 6 |
| 47-kDa antigen | PGN_0852 | 7.8E+06 | 0 | 4.1E+05 | 4 | |
| Transketolase | PGN_1689 | 5.5E+06 | 0 | 2.1E+05 | 2 | |
| CPG70 | PGN_0335 | 4.2E+06 | 0 | 1.2E+05 | 2 | |
| Ribonuclease E/G | PGN_1391 | 4.0E+06 | 0 | 8.3E+04 | 3 | |
| AcrB-type transporter | PGN_1430 | 5.2E+06 | 0 | 7.6E+04 | 2 | |
T9SS cargo proteins are shaded.
RgpB is inferred considering that peptides in common between RgpA and RgpB were enriched in the mix, whereas the RgpA-specific peptides were depleted (see Materials and Methods and Fig. 5). These data were therefore produced by removing the RgpA sequence from the database.
Because the sequences of RgpA and RgpB catalytic domains are very similar, the data generated using both sequences in the database could not distinguish between the two proteins.
The locus tag can be found in the National Center for Biotechnology Information database (NCBI), https://www.ncbi.nlm.nih.gov.
FIG 5.
MaxQuant data for RgpA/RgpB peptides identified in the anti-A-LPS co-IP. The intensity data for the 9 common peptides identified for RgpA/RgpB are compared to those for the 3 RgpA-specific peptides. For each peptide, the intensity in the complemented (mix) sample is compared to the porU negative control (since the wzy control was of lower abundance). The ratio of each pair is provided above the columns.
Additionally, since the sequence of RgpB is different between the two strains, the dominant sequence detected in the mixed sample should indicate which strain was providing the RgpB for modification. Sequence comparison predicts four sequence differences that should produce two sets of different tryptic peptides (see Table S1 and Fig. S2 in the supplemental material). Searching the data against both strain databases (33277 and 381) revealed that only the 33277 variant peptides (1 and 9) (Fig. 5; also, see Table S1) were identified, consistent with the RgpB detected being produced by the 33277 wzy strain. These 2 peptides are common to RgpA of both strains (Table S1); however, it is highly unlikely that they were from RgpA, since these peptides were, respectively, 10-fold less abundant and undetected in the 381 porU control strain (Fig. 5) and even less abundant in the 33277 wzy control strain (data not shown).
Rhodamine-labeled T9SS mutant OMVs fuse with A-LPS biosynthetic mutant cells.
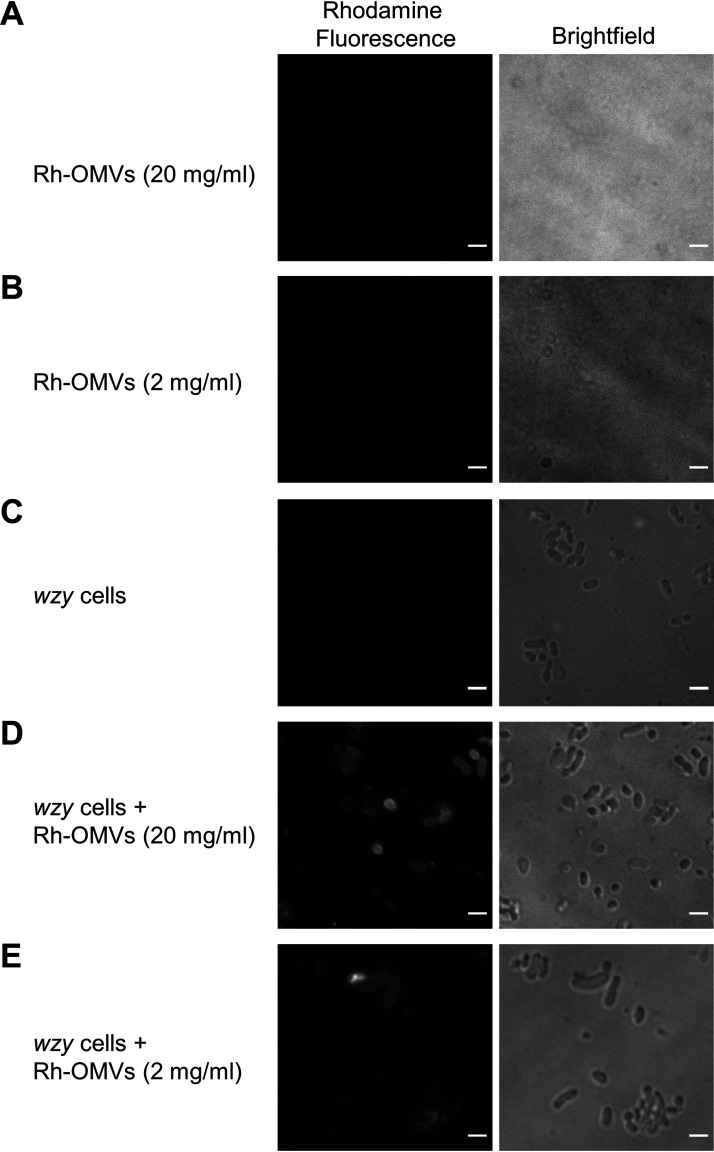
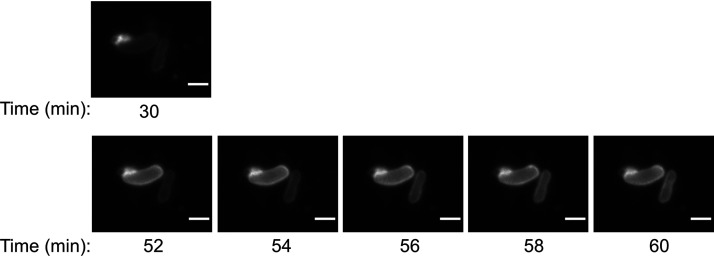
The fluorescence of rhodamine B-R18 in membranes is quenched at high dye concentrations but is released upon dilution. This property makes the dye useful for membrane fusion assays. To provide evidence that lipid-associated molecules can be exchanged between OMVs and P. gingivalis cells via membrane fusion, the rhodamine OMV fusion assay (51) was employed. The OMVs from the porU mutant were labeled with rhodamine B-R18 and mixed at two different concentrations with wzy mutant cells. The labeled OMV membranes and wzy cells did not fluoresce (Fig. 6A to C), but when the labeled OMVs were mixed with wzy cells, the cell membranes of a proportion of cells could be observed fluorescing, indicating that an exchange of rhodamine dye had occurred between the OMVs and cells (Fig. 6D and E). Figure 7 captures this fusion and dye transfer process in action, showing fluorescence at the contact region between the rhodamine B-R18-labeled OMVs (Rh-OMVs) and one cell and then presenting a time course beginning 22 min later, showing the transfer of dye to a second cell.
FIG 6.
Rhodamine-stained OMVs transfer lipophilic dye to 33277 wzy cells. (A and B) Rhodamine-labeled OMVs from the 381 porU mutant (Rh-OMVs) shown at two different concentrations: 20 mg/ml (A) and 2 mg/ml (B). (C) 33277 wzy-only cells (negative-control cells). (D) 33277 wzy cells mixed with Rh-OMVs at 20 mg/ml. (E) 33277 wzy cells mixed with Rh-OMVs at 2 mg/ml. A single Z slice is presented. Bars, 2 μm.
FIG 7.
Live capture of OMV-to-cell transfer of rhodamine B-R18 lipid dye. (Top) Close-up of two cells shown in Fig. 6E (30 min after mixing of wzy cells and rhodamine-labeled porU OMVs). (Bottom) Same cells showing a time-lapse series of rhodamine fluorescence every 2 min for 8 min. A single Z slice is presented. Bars, 2 μm.
A-LPS from porU OMVs binds to the OM of wzy cells.
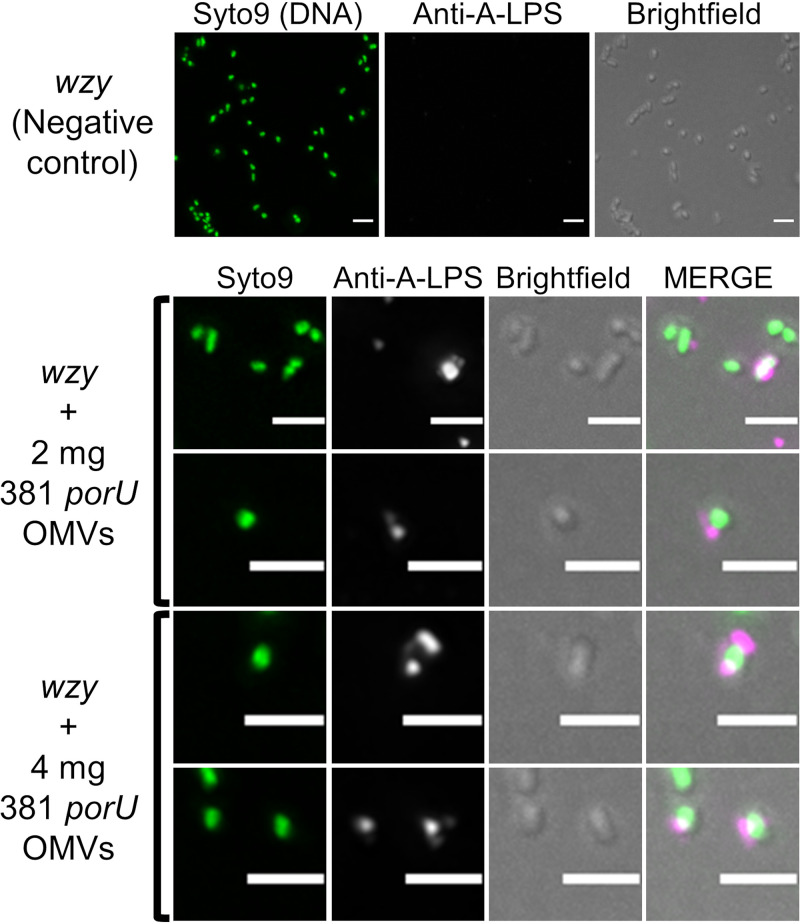
To observe fusion of A-LPS to the OM of the recipient cell, OMVs from the porU mutant were mixed with wzy cells and incubated on BHI-T BA for 4 days. Cells were scraped into phosphate-buffered saline (PBS) and fluorescently stained with anti-A-LPS. Fluorescently stained wzy-only control cells were negative, while 18% of cells mixed with 2 mg of OMVs stained positive to various degrees (over 11,000 cells counted), and 26% of cells were positive when mixed with 4 mg of OMVs (over 7,000 cells counted) (Fig. 8). Most positive cells showed one or more patches of fluorescence in proximity to their surface, indicating that OMVs had adhered to the cells (Fig. 8); however, higher resolution is required to show fusion of the OMVs with the OM or transfer of A-LPS from the OMV to the OM of the cell.
FIG 8.
Free A-LPS from porU OMVs binds to the surfaces of wzy cells. Negative-control wzy cells only or wzy cells mixed with 2 mg or a total of 4 mg of purified OMVs from 381 porU were spread onto BHI-T BA plates and incubated anaerobically at 37°C for 5 days. Cells were scraped into PBS, spun, and resuspended to the same OD650. Cells were blocked, fluorescently stained by incubation with anti-A-LPS, washed, incubated with AF647-conjugated goat anti-mouse IgG, and then washed again. A maximum-intensity projection of all Z stacks is presented. Bars, 2 μm.
DISCUSSION
To better understand how the OM components of the T9SS machinery function, we wanted to investigate whether they could be complemented in trans. We initially used all combinations of a two-mutant-strain crossover grown on blood agar to look for reacquisition of black pigmentation within the mixed population. This analysis focused on five of the T9SS OM components (encoded by porU, porV, porT, porQ, and porZ) and two A-LPS biosynthetic mutants (wbaP and wzy mutants). None of the two-strain crosses involving only the five T9SS component mutants produced black pigmentation, implying that the OM proteins cannot be transferred in a manner that restores function and that, hence, a more ordered temporal assembly may be required. It could not be distinguished whether the OM proteins were not transferred or whether they were transferred but not able to be delivered or incorporated into the T9SS. If OMV fusion occurred, both OM proteins as well as periplasmic proteins could potentially be transferred, which is relevant for PorU and PorZ, since they are located in the periplasm of most T9SS mutants. The proteins tested (PorU, PorV, PorQ, PorZ, and PorT) are among the simplest components to be transferred, as many of the other OM components (Sov, PorW, PorE, PorK, PorN, and PorG) are part of larger assemblies or are bound to the cell wall (39, 41, 52). Therefore, the results strongly suggest that T9SS components cannot be exchanged between cells to compensate for mutations in the two strains mixed.
Of all the two-strain crosses, only those involving an A-LPS biosynthetic mutant, i.e., a wbaP or wzy mutant, with a T9SS component mutant produced black pigmentation and therefore complemented the A-LPS biosynthesis mutation. Western blot analyses of the cell lysates from these crosses involving wzy or wbaP confirmed that A-LPS was able to be covalently linked to cargo proteins (Fig. 2). The size of these A-LPS-modified cargo proteins matched the size of the WT strain corresponding to the T9SS mutant, indicating that in each case, the T9SS mutant was the A-LPS donor. Assuming a donor-recipient model (see below), this supports the idea that A-LPS is directly utilized from the donor and not remetabolized by the recipient strains (wzy or wbaP).
After complementation between 33277 wzy and 381 porU mutants, most of the proteins associated with A-LPS, as identified by co-IP using anti-A-LPS, were cargo proteins (Table 1). This further confirmed that complementation involved the A-LPS modification of cargo proteins. It is not known whether the other noncargo proteins identified as being associated with A-LPS play a role in conjugation of A-LPS, but they are not expected to be localized to the OM (15, 53) and probably bind artefactually to A-LPS after cell lysis. The A-LPS-associated cargo proteins observed from co-IP capture tended to be the most abundant cargo proteins in P. gingivalis according to MaxQuant analyses of whole-cell culture lysates (54); however, the abundant gingipains Kgp and RgpA were missing. Both Kgp and RgpA form very large complexes of >450 kDa (16). The lack of their detection by co-IP could therefore have been due to steric hindrance. The co-IP evidence supports the Western blot analyses for the same mutant cross (Fig. 2C and E), showing that the cargo proteins are covalently linked to A-LPS when complementation is observed.
Complementation involving A-LPS conjugation of cargo proteins could occur via two distinct pathways. One could envisage a functional attachment complex on the surface of the A-LPS mutant catalyzing the “intercell” conjugation of A-LPS to cargo proteins located on the T9SS mutant. Alternatively, as we propose, the A-LPS must transfer from the donor T9SS mutant to the OM of the recipient A-LPS mutant and then be conjugated to the cargo proteins of the recipient strain in the normal way. We have demonstrated the latter pathway in two ways. The RgpB present in the co-IP was identified to be from the recipient strain (33277 wzy) and furthermore, the wzy mutant could also be complemented by providing purified P. gingivalis LPS in trans (from an exogenous source without cargo proteins). The mechanism of complementation proposed involves the donation of A-LPS from any source (cells, OMVs, or purified LPS) to a cell originally lacking A-LPS but having a functional T9SS (Fig. 9). These results therefore also show that the WbaP and Wzy enzymes are required only for production of A-LPS and not for its conjugation, since the mutants lacking these enzymes require only addition of free A-LPS for complementation of the biosynthesis defect.
FIG 9.
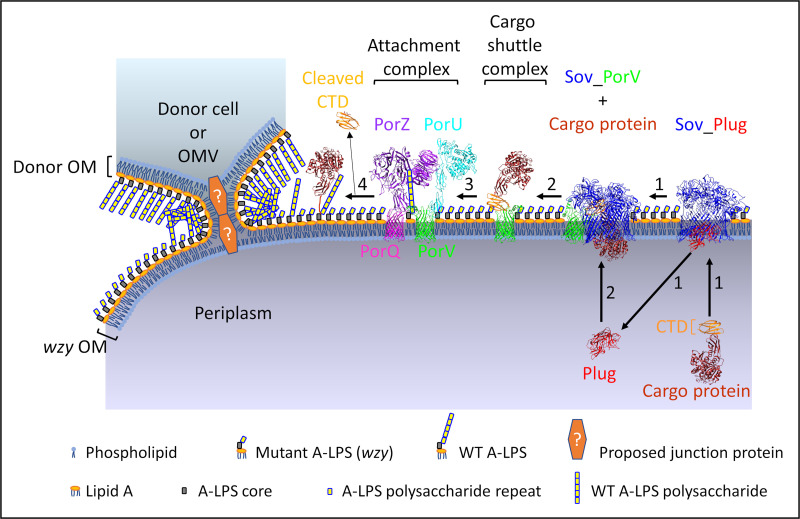
Proposed model for LPS exchange and A-LPS complementation in P. gingivalis. Proposed model for complementation of the wzy LPS mutant by LPS exchange with donor cell or OMV possessing WT unconjugated A-LPS. The OM of wzy cell fuses with the OM of the donor cell or OMV by forming a tight junction via an unknown OM adhesin or junction protein. Donor WT A-LPS then transfers to the OM of wzy cell to provide functional A-LPS for the T9SS attachment complex. Step 1, PorV (green) plugs the side opening of the Sov (blue) pore, and the plug protein (red) exits from the pore, allowing the cargo protein to enter the Sov pore and interact with the PorV loops that are inside the Sov channel (39). Step 2, the cargo shuttle complex comprising PorV bound to the CTD (orange) of the cargo protein (brown) is released from Sov, and the plug protein reenters from the periplasm. Step 3, the cargo shuttle complex interacts with the attachment complex (PorV-PorU-PorQ-PorZ) that is primed via PorZ (purple) with WT A-LPS derived from the donor cell/OMV, allowing the CTD of the cargo protein to be cleaved by PorU (cyan) and the new C terminus of the cargo protein to be conjugated to the WT A-LPS for anchorage to the OM and release of cleaved CTD (step 4) (7, 8, 38). If WT A-LPS is not present, the CTD of the cargo protein is still cleaved by PorU but is not conjugated to A-LPS, and it is released from the cell surface. The second LPS of P. gingivalis, O-LPS, is not represented. Structures for Sov_Plug (PDB ID: 6H3J) and Sov_PorV (PDB ID: 6H3I) complexes were determined by cryo-electron microscopy (39). The crystal structures for mature RgpB (PDB ID: 1CVR) (90) and RgpB CTD (PDB ID: 5AG9) (91) were used for the cargo protein. PorZ is from the crystal structure (PDB ID: 5m11) (43). PorV (green) and PorQ (magenta) β-barrel structures are from Phyre2 models using amino acid sequences without N-terminal signal peptides. The PorU structure is a Phyre2 model without a signal peptide or domain A (7). Attachment and cargo shuttle complexes are depicted based on composition of complexes and subcomplexes (16, 38).
It was shown previously that OMVs deliver periplasmic proteins and LPS between Gram-negative bacteria due to binding/fusion of OMVs to bacterial cells (46, 49). LPS exchange, indirectly by functional complementation, and direct lipoprotein exchange have been elegantly shown in the multicellular Gram-negative organism Myxococcus xanthus; this process is known as OM exchange (50). This process is dependent on the cell surface receptor TraA and the TraB cohort protein due to TraA binding between cells to form a tight junction for LPS exchange to occur. In this species, LPS O antigen is required for development (55) and for cells to move as groups in a biofilm powered by type 4 pilus extension and retraction, known as social gliding motility (56). OM exchange was observed only when cells were grown on a solid surface and did not occur in liquid medium even when TraAB were overexpressed (50). Likewise, complementation of the A-LPS biosynthetic mutants occurred only when cells were grown on solid medium and did not occur in liquid medium (data not shown). TraAB homologues are absent in P. gingivalis, so LPS exchange in this species is likely to occur via a novel contact-dependent mechanism.
We have used the method of Bomberger et al. (51) to show the exchange of the special fluorescent lipid dye, rhodamine B-R18, between OMVs and P. gingivalis cells. At high concentrations in bilayer membranes, fluorescence of this dye is quenched, but when the probe is diluted upon OMV fusion with unlabeled membranes, fluorescence is “dequenched,” allowing fluorescence to be observed. Fluorescence of this lipid dye was observed after rhodamine-labeled porU OMVs were incubated with wzy cells (Fig. 6D and E). This lipid dye transfer was also captured during imaging (Fig. 7). Immunofluorescence labeling of A-LPS using anti-A-LPS for the mix of wzy cells and porU OMVs showed A-LPS in OMVs bound to the surface of cells (Fig. 8), which increased with an increase in the amount of OMVs added. Together with the observation of complementation using different sources of A-LPS with A-LPS biosynthetic mutants, these results demonstrate that exchange of LPS between P. gingivalis cells is possible and that exchange between P. gingivalis and other Gram-negative bacteria may also be possible (4).
It is well established that the acquisition of iron plays an important role in microbial virulence (57, 58) and is particularly relevant for P. gingivalis (59, 60). P. gingivalis obtains its iron from hemin, but as P. gingivalis is auxotrophic for heme (61), hemin is an absolute requirement for growth (62–64). The availability of iron in the host is extremely low due to strong sequestration by host hemin-binding proteins, such as hemoglobin-haptoglobin complex, hemopexin, and albumin; therefore, P. gingivalis has evolved very efficient systems for the extraction, capture, and transportation of heme (30, 44, 65, 66). P. gingivalis is also very efficient at the storage of heme on the cell surface in the form of a black-pigmented heme dimer, μ-oxo bisheme, through binding to A-LPS and hemin-binding receptors, such as certain T9SS cargo proteins (14, 44, 65, 66). P. gingivalis has a 10-fold-greater specific hemin binding affinity and greater heme storage capacity than Prevotella intermedia, a less severe periodontitis-associated species belonging to the orange complex (67, 68). The ability of P. gingivalis to store the heme dimer has relevance to this study, since it has the potential to be transferred, together with A-LPS, to other bacteria through close contact or through fusion with P. gingivalis OMVs. P. gingivalis may therefore affect the availability of iron for other oral periodontitis-relevant bacterial species that require hemin for growth, such as those belonging to the red complex, which is associated with the most severe pathology (P. gingivalis, Tannerella forsythia, and Treponema denticola; the latter two species are not black pigmented) (68) and which can form a polymicrobial biofilm (69). Other polymicrobial communities are known to affect iron availability in the oral cavity (70). The possibility of LPS-hemin exchange with P. gingivalis may contribute to an important polymicrobial synergy in the development of periodontitis.
The potential to exchange LPS between oral bacteria has other important implications in the development of periodontitis. P. gingivalis requires other bacteria (oral microbiota) and chronic inflammation to emerge and cause disease (71). This study is the first known example of LPS exchange to be shown in a human bacterial species whose pathogenicity depends on complex interactions with other bacterial species. Exchange of LPS with other bacterial species would be expected to change how P. gingivalis interacts with the host through acquiring changes in LPS structure (72, 73), influencing complement activation and host cell invasion by the pathogen (74) or internal activation of pyroptosis after epithelial invasion (75–84).
In conclusion, we have demonstrated that important OM components of the T9SS cannot be exchanged between mutants to complement their genetic defects, implying that a more ordered temporal assembly of the T9SS is required. We have also demonstrated, using a multiassay approach, the ability of P. gingivalis to exchange A-LPS between strains, raising the possibility that LPS exchange also occurs with other Gram-negative bacteria within the oral microbiota, which would be expected to exert diverse effects on the microbial community and its interactions with the human host.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Porphyromonas gingivalis WT strains W50 (ATCC 53978) and 381 were from the Oral Health Cooperative Research Centre, University of Melbourne, Melbourne, Australia. ATCC 33277 and the 33277 porQ mutant were described previously (9) and were a kind gift from Koji Nakayama. Previously described isogenic mutants are W50 porV (lptO) and W50 porT (37); W50 porU (PG26) (7); W50 wbaP (8); W50 porZ (38); 33277 porT (33); and 33277 wzy (14). The porV, porU, porUC690A, and porZ mutants were constructed in the 381 strain background using plasmids and methods previously described (7, 37, 38). Previously, these strains were thought to be in the 33277 strain background; however, resequencing of the strain demonstrated a closer match to 381 (data not shown).
P. gingivalis cells were grown on several growth media at 37°C under anaerobic conditions (80% N2, 10% CO2, 10% H2). Trypticase soy and bovine heart infusion (TSBHI) broth (37) was supplemented with 0.5 mg/ml cysteine, 5 μg/ml hemin, and 5 μg/ml vitamin K. BHI-T BA is TSBHI agar (7) supplemented with 0.5 mg/ml cysteine, 5 μg/ml hemin, 5 μg/ml vitamin K, and 5% (vol/vol) horse blood. BHI-T is BHI-T BA without horse blood. BBA is 40 g/liter of blood agar base (Oxoid) with 7% (vol/vol) horse blood and 5 μg/ml vitamin K. BHI broth is 37 g/liter of bovine heart infusion (Becton Dickinson and Company) supplemented with 0.5 mg/ml cysteine, 5 μg/ml hemin, and 5 μg/ml vitamin K.
Complementation on plates.
The mixing of two mutant P. gingivalis strains to test for reacquisition of T9SS functionality by observation of black pigmentation was performed in two ways. The first method applied two mutant strains from glycerol stocks onto BHI-T BA medium. The first strain was applied in a vertical line, and the second strain was applied in one horizontal motion from the left to the right through the first strain. The plates were then incubated anaerobically for 14 days at 37°C. The second method involved mixing two mutant strains from 6 μl of glycerol stock each on BHI-T BA medium using a sterile loop to form a small oval patch and incubating plates anaerobically for 6 days at 37°C. The single strains were patched on either side of the mixed patch using the same volume of glycerol stocks. After growth, cells in patches were then scraped off plates into 1 ml of PBS, resuspended, pelleted by centrifugation at 8,000 × g for 20 min at 4°C and stored at −80°C.
For testing purified P. gingivalis LPS in a complementation assay, glycerol stock of the 33277 wzy strain was diluted either 1:4 or 1:9 with TSBHI broth, containing cysteine, hemin, and vitamin K additives as described above, and spread onto BHI-T BA medium to make a lawn of cells. Small sterile square blotting paper pieces were then placed onto the lawn, and 20-μl volumes of ultrapurified LPS from the ATCC 33277 strain of P. gingivalis (InvivoGen) were applied separately at 1 μg/μl (reconstituted in sterile water provided) and 10-fold dilutions in sterile water. The plates were then incubated anaerobically for 15 days at 37°C.
For testing purified OMVs in a complementation assay, 10 μl of 33277 wzy glycerol stock diluted 1:4 with BHI-T broth (for plating on BHI-T medium) or BHI broth (for plating on BBA medium) was mixed with 10 μl of sterile water (control) or with 10 μl of purified 381 porU OMVs resuspended in sterile water at 200 μg/μl (M1), 100 μg/μl (M2), 50 μg/μl (M3), or 25 μg/μl (M4) and incubated at 37°C for 5 min before being spread into small circular patches on BHI-T BA or BBA plates and incubated anaerobically for 4 days at 37°C. Cells in patches were then scraped off plates into 1 ml of PBS, resuspended, and pelleted by centrifugation at 8,000 × g for 20 min at 4°C, the supernatant was removed, and the cell pellet was at −80°C.
Purification of OMVs.
OMVs were isolated from the filtered culture supernatant from strains grown to late log phase as previously described (16), except that OMV pellets were not washed in PBS. OMV pellets were resuspended to a wet weight of 200 mg/ml in sterile Milli-Q water before use.
SDS-PAGE and Western blotting.
For whole-cell lysates, cells were lysed in lysis buffer containing 0.5% (vol/vol) Triton X-114, 25 mM Tris-HCl (pH 7.4), 2× lithium dodecyl sulfate (LDS) sample buffer, 20 mM Nα-tosyl-l-lysine chloromethyl ketone hydrochloride (TLCK; Sigma-Aldrich), a 1/50 dilution of protease inhibitor cocktail (Sigma-Aldrich), and 50 mM dithiothreitol and then boiled 4 min before loading the SDS-PAGE gel. Lysates from cell pellets were normalized to a wet weight per milliliter. SDS-PAGE was performed using NuPAGE Novex bis-Tris 4 to 12% or 10% gels (1 mm thick; 15 wells or 26 wells) and MOPS (morpholinepropanesulfonic acid) running buffer. The SDS-PAGE gel was then transferred to nitrocellulose in 1× transfer buffer (Life Technologies) with 10% (vol/vol) methanol. The membrane was stained with Ponceau S, destained with PBS containing 0.05% (vol/vol) Tween 20 (PBST), blocked for 1 h at room temperature in PBST containing either 1% (wt/vol) bovine serum albumin (BSA) or 5% (wt/vol) skim milk, incubated with diluted specific antibody overnight at room temperature, washed three times in PBST, incubated with secondary antibody (goat anti-mouse IgG conjugated to horseradish peroxidase [Sigma-Aldrich] diluted 1/2,000 in 5% [wt/vol] skim milk in PBST), incubated for 1 h at room temperature, washed three times again, and then developed using SuperSignal West Pico Plus chemiluminescent substrate (Thermo Fisher Scientific). Hybridoma culture supernatant containing mouse monoclonal antibody (MAb) 1B5, also called anti-A-LPS, recognizes an epitope found on A-LPS (17) and was a gift from M. A. Curtis. Anti-A-LPS was used at a dilution of 1/100 in 1% (wt/vol) BSA in PBS. Anti-CPG70 (85) was used at a dilution of 1/1,000 in 5% (wt/vol) skim milk in PBST.
Anti-A-LPS whole-cell ELISA.
Whole-cell ELISA was performed as previously described (38). Briefly, P. gingivalis strains were grown on BHI-T with the appropriate antibiotic and passaged on BHI-T without selection for 4 days. To make formaldehyde-killed whole cells (FKWCs), cells were scraped off plates, washed twice in 50 mM Tris (pH 7.4), 150 mM NaCl, and 5 mM CaCl2, and treated with formalin saline (0.5% [vol/vol] formaldehyde in 8.5 g/liter NaCl) overnight at room temperature with gentle inversion, followed by two washes in PBS. FKWCs were added (108 per well) to a Corning 96-well flat-bottom polystyrene high-binding microplate and incubated overnight at 4°C. The solution was aspirated, and wells were blocked with 1% (wt/vol) BSA in PBS. Anti-A-LPS was 2-fold serially diluted in blocking solution starting at 1/100; 50 μl was added per well in triplicate and incubated for 2.5 h at room temperature. The plates were then washed 3 times in PBST, and 50 μl per well of goat anti-mouse IgG antibody conjugated to horseradish peroxidase diluted 1/1,000 in blocking solution was added and incubated for 2 h at room temperature. The plates were then washed 3 times and developed with the peroxidase substrate 2,2′-azino-bis(3-ethylbenzothiazoline 6-sulfonic acid) diammonium salt. When optimum color development had been reached, the reaction was stopped by addition of NaF, and optical density (OD) was measured at 405 nm.
MaxQuant analysis of coimmunoprecipitated samples.
Cell lysates were obtained after growing 33277 wzy with 381 porU mutants together (mix) and separately on BHI-T BA for 6 days and were subjected to co-IP using anti-A-LPS. Equivalent amounts of cell pellets were resuspended in 20 mM Tris-HCl (pH 8), 100 mM NaCl, 1% n-dodecyl-β-d-maltoside (DDM), 5 mM TLCK and complete protease inhibitors and incubated on ice for 45 min. The lysates were clarified by centrifugation at 16,000 × g for 10 min at 4°C. Cell lysates were mixed with anti-A-LPS antibodies bound to agarose (Sigma-Aldrich). The beads were rotated on a wheel for 2 h at 4°C and then washed with 20 mM Tris-HCl (pH 8), 100 mM NaCl, followed by one high-salt wash with 20 mM Tris-HCl (pH 8), 500 mM NaCl and lastly with 10 mM Tris-HCl (pH 8). The beads were resuspended in Laemmli buffer and boiled for 10 min. The samples were subjected to SDS-PAGE for a short time and stained with Coomassie blue. Each sample was excised as a single band and subjected to tryptic digestion and LC-MS/MS analysis as previously described (54).
The MS raw files were analyzed by MaxQuant (version 1.5.3.30) (86) against the ATCC 33277 P. gingivalis sequence database with the default recommended parameters except that the “match between runs” and iBAQ options were activated and the acetylation modification was removed. The settings included a minimal requirement of two peptides to be quantitated. The protein data were found in the proteinGroups.txt file in the MaxQuant output. To determine which proteins were specifically immunoprecipitated, the LFQ intensity ratio between the mix and the higher of the two negative controls was calculated. Proteins with LFQ ratios of >3 and proteins that were detected only in the mix were deemed significant. The relative abundance of these proteins was calculated as the iBAQ intensity minus the iBAQ intensity from the higher of the two negative controls. To enable quantitation of RgpB, the MaxQuant analysis was repeated with the RgpA sequence removed from the database. The intensities of individual RgpA and RgpB peptides were found in the peptides.txt file in the MaxQuant output. The MS data were also searched against the sequence database of strain 381; however, no new peptides of interest (from T9SS cargo proteins) were identified.
OMV-cell fusion and fluorescence microscopy.
Purified OMVs were washed twice in 140 mM NaCl, 12 mM K2HPO4, 14 mM Na2HPO4, 25 mM glucose (pH 7.5) (Pg-PBS buffer) at 170,000 × g for 1 h at 8°C. The OMVs were fluorescently labeled with octadecyl rhodamine B chloride (rhodamine B-R18) (Thermo Fisher Scientific), a probe that fluoresces only upon membrane fusion due to a dilution-based dequenching effect, according to the method of Bomberger et al. (51). Briefly, 0.05 g of washed OMVs was resuspended in 1 ml of labeling buffer (50 mM Na2CO3, 100 mM NaCl [pH 9.2]) containing 1 mg/ml of rhodamine B-R18 and incubated for 1 h at 25°C, followed by two washes in 37 ml of Pg-PBS and finally resuspended to 200 mg/ml (wet weight) in Pg-PBS. Freshly grown mid-log-phase wzy cells (1 ml) were centrifuged at 10,000 × g for 5 min at 18°C and resuspended in 100 μl of culture medium (TSBHI broth with cysteine, hemin, and vitamin K additives) containing 20 mg/ml of Rh-OMVs. The mix of cells and Rh-OMVs were incubated at 37°C for 30 min, then washed twice in 1 ml of Pg-PBS at 10,000 × g for 5 min at 18°C, and resuspended in 100 μl of Pg-PBS. Alternatively, 1 ml of mid-log-phase wzy cells were centrifuged at 10,000 × g for 5 min at 18°C, resuspended in 100 μl of Pg-PBS containing 2 mg/ml of Rh-OMVs, and then incubated for 30 min at 37°C. Rh-OMVs only were added to either culture medium at 20 mg/ml or Pg-PBS at 2 mg/ml and incubated for 30 min at 37°C. Z-stacks of images were acquired with a DV Elite microscope (Olympus U-Plan S-Apo 100×/1.42 numerical aperture [NA] objective and SoftWoRx 6.1 software [GE Healthcare]) with heating at 37°C, using excitation at 575 nm and emission at 632 nm for rhodamine fluorescence and immersion oil with a refractive index of 1.518. Image files were analyzed using ImageJ software (National Institutes of Health). Experiments were repeated twice with 2 to 6 representative fields imaged for each sample.
For immunostaining of cells with anti-A-LPS, negative-control wzy cells only or wzy cells mixed with 2 mg of purified OMVs from 381 porU were spread onto BHI-T BA medium and incubated anaerobically at 37°C for 5 days. After 3 days’ incubation on BHI-T BA, a duplicate mix of wzy cells with 2 mg of porU OMVs was mixed with another 2 mg of porU OMVs and incubated a further 2 days (4 mg [total amount] of porU OMVs added). Cells were scraped into PBS, spun, and resuspended to the same optical density at 650 nm (OD650). Cells were blocked with 3% (wt/vol) BSA in Pg-PBS containing 10 mM TLCK for 1 h at 4°C, fluorescently stained by incubation with anti-A-LPS, washed twice in Pg-PBS, incubated with Alexa Fluor 647-conjugated goat anti-mouse IgG (Thermo Fisher Scientific) for 35 min at 4°C, and washed again. Immunostained cells were adhered to poly-l-lysine-treated coverslips overnight at 4°C, washed twice with Pg-PBS, and stained with Syto9 (Life Technologies). Images were captured on a DeltaVision Elite microscope (Olympus U-Plan S-Apo 100×/1.42 NA objective; excitation at 475 nm and emission at 523 nm for Syto9; excitation at 632 nm and emission at 676 nm for Alexa Fluor 647), deconvolved using softWoRx 6.1 software, and analyzed using ImageJ software.
Generation of protein structure graphics.
Molecular graphics were produced using UCSF Chimera version 1.14 software (http://www.cgl.ucsf.edu/chimera) (87) using structural data retrieved from the Protein Data Bank (PDB; http://www.wwpdb.org) (88). Protein structure models were produced for proteins with unsolved structures using Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2) (89).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Australian Government Department of Industry, Innovation and Science (grant ID 20080108) and the National Health and Medical Research Council of Australia (grant ID 1123866).
We thank Nuzul Nur Iman for his help in producing Fig. 3. We thank Yu-Yen Chen for providing the mouse anti-CPG70 serum. We acknowledge the use of the Mass Spectrometry and Proteomics Facility and the Biological Optical Microscopy Platform, both at the Bio21 Institute, The University of Melbourne, Australia.
Michelle D. Glew: Conceptualization, Data curation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing. Dhana G. Gorasia: Data curation, Formal analysis, Writing - review & editing. Paul J. McMillan: Data curation, Investigation, Writing - review & editing. Catherine A. Butler: Formal analysis, Writing - review & editing. Paul D. Veith: Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Writing - review & editing. Eric C. Reynolds: Funding acquisition, Supervision, Resources, Project administration, Writing - review & editing.
We declare that we have no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 8:481–490. 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 2.Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat Rev Microbiol 10:717–725. 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 15:30–44. 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirst ME, Li EC, Alfant B, Chi YY, Walker C, Magnusson I, Wang GP. 2015. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl Environ Microbiol 81:783–793. 10.1128/AEM.02712-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O-Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. 2003. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Pept Sci 4:409–426. 10.2174/1389203033487009. [DOI] [PubMed] [Google Scholar]
- 6.Fitzpatrick RE, Wijeyewickrema LC, Pike RN. 2009. The gingipains: scissors and glue of the periodontal pathogen, Porphyromonas gingivalis. Future Microbiol 4:471–487. 10.2217/fmb.09.18. [DOI] [PubMed] [Google Scholar]
- 7.Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds EC. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem 287:24605–24617. 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorasia DG, Veith PD, Chen D, Seers CA, Mitchell HA, Chen YY, Glew MD, Dashper SG, Reynolds EC. 2015. Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog 11:e1005152. 10.1371/journal.ppat.1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to Bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107:276–281. 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J Bacteriol 195:270–278. 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakayama K. 2015. Porphyromonas gingivalis and related bacteria: from colonial pigmentation to the type IX secretion system and gliding motility. J Periodontal Res 50:1–8. 10.1111/jre.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasica AM, Ksiazek M, Madej M, Potempa J. 2017. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol 7:215. 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veith PD, Glew MD, Gorasia DG, Reynolds EC. 2017. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol 106:35–53. 10.1111/mmi.13752. [DOI] [PubMed] [Google Scholar]
- 14.Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. 2011. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6:e21372. 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veith PD, Chen YY, Gorasia DG, Chen D, Glew MD, O'Brien-Simpson NM, Cecil JD, Holden JA, Reynolds EC. 2014. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res 13:2420–2432. 10.1021/pr401227e. [DOI] [PubMed] [Google Scholar]
- 16.Glew MD, Veith PD, Chen D, Seers CA, Chen YY, Reynolds EC. 2014. Blue native-PAGE analysis of membrane protein complexes in Porphyromonas gingivalis. J Proteomics 110:72–92. 10.1016/j.jprot.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 17.Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, Hounsell E, Curtis MA. 2005. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol Microbiol 58:847–863. 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- 18.Paramonov N, Aduse-Opoku J, Hashim A, Rangarajan M, Curtis MA. 2015. Identification of the linkage between A-polysaccharide and the core in the A-lipopolysaccharide of Porphyromonas gingivalis W50. J Bacteriol 197:1735–1746. 10.1128/JB.02562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen T, Dong H, Yong R, Duncan MJ. 2000. Pleiotropic pigmentation mutants of Porphyromonas gingivalis. Microb Pathog 28:235–247. 10.1006/mpat.1999.0338. [DOI] [PubMed] [Google Scholar]
- 20.Shoji M, Yukitake H, Sato K, Shibata Y, Naito M, Aduse-Opoku J, Abiko Y, Curtis MA, Nakayama K. 2013. Identification of an O-antigen chain length regulator, WzzP, in Porphyromonas gingivalis. Microbiologyopen 2:383–401. 10.1002/mbo3.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanterpool E, Roy F, Fletcher HM. 2005. Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect Immun 73:3971–3982. 10.1128/IAI.73.7.3971-3982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi M, Sato K, Yukitake H, Noiri Y, Ebisu S, Nakayama K. 2010. A Porphyromonas gingivalis mutant defective in a putative glycosyltransferase exhibits defective biosynthesis of the polysaccharide portions of lipopolysaccharide, decreased gingipain activities, strong autoaggregation, and increased biofilm formation. Infect Immun 78:3801–3812. 10.1128/IAI.00071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein BA, Cornacchione LP, Collins M, Malamy MH, Duncan MJ, Hu LT. 2017. Using Tn-seq to identify pigmentation-related genes of Porphyromonas gingivalis: characterization of the role of a putative glycosyltransferase. J Bacteriol 199:e00832-16. 10.1128/JB.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoji M, Sato K, Yukitake H, Kamaguchi A, Sasaki Y, Naito M, Nakayama K. 2018. Identification of genes encoding glycosyltransferases involved in lipopolysaccharide synthesis in Porphyromonas gingivalis. Mol Oral Microbiol 33:68–80. 10.1111/omi.12200. [DOI] [PubMed] [Google Scholar]
- 25.Veith PD, Shoji M, O’Hair RAJ, Leeming MG, Nie S, Glew MD, Reid GE, Nakayama K, Reynolds EC. 2020. Type IX secretion system cargo proteins are glycosylated at the C terminus with a novel linking sugar of the Wbp/Vim pathway. mBio 11:e01497-20. 10.1128/mBio.01497-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoji M, Sato K, Yukitake H, Naito M, Nakayama K. 2014. Involvement of the Wbp pathway in the biosynthesis of Porphyromonas gingivalis lipopolysaccharide with anionic polysaccharide. Sci Rep 4:5056. 10.1038/srep05056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abaibou H, Chen Z, Olango GJ, Liu Y, Edwards J, Fletcher HM. 2001. vimA gene downstream of recA is involved in virulence modulation in Porphyromonas gingivalis W83. Infect Immun 69:325–335. 10.1128/IAI.69.1.325-335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanterpool E, Roy F, Sandberg L, Fletcher HM. 2005. Altered gingipain maturation in vimA- and vimE-defective isogenic mutants of Porphyromonas gingivalis. Infect Immun 73:1357–1366. 10.1128/IAI.73.3.1357-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato K, Kido N, Murakami Y, Hoover CI, Nakayama K, Yoshimura F. 2009. Lipopolysaccharide biosynthesis-related genes are required for colony pigmentation of Porphyromonas gingivalis. Microbiology (Reading) 155:1282–1293. 10.1099/mic.0.025163-0. [DOI] [PubMed] [Google Scholar]
- 30.Rangarajan M, Aduse-Opoku J, Paramonov NA, Hashim A, Curtis MA. 2017. Hemin binding by Porphyromonas gingivalis strains is dependent on the presence of A-LPS. Mol Oral Microbiol 32:365–374. 10.1111/omi.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dou Y, Osbourne D, McKenzie R, Fletcher HM. 2010. Involvement of extracytoplasmic function sigma factors in virulence regulation in Porphyromonas gingivalis W83. FEMS Microbiol Lett 312:24–32. 10.1111/j.1574-6968.2010.02093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadowaki T, Yukitake H, Naito M, Sato K, Kikuchi Y, Kondo Y, Shoji M, Nakayama K. 2016. A two-component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Sci Rep 6:23288. 10.1038/srep23288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato K, Sakai E, Veith PD, Shoji M, Kikuchi Y, Yukitake H, Ohara N, Naito M, Okamoto K, Reynolds EC, Nakayama K. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J Biol Chem 280:8668–8677. 10.1074/jbc.M413544200. [DOI] [PubMed] [Google Scholar]
- 34.Ishiguro I, Saiki K, Konishi K. 2009. PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system. FEMS Microbiol Lett 292:261–267. 10.1111/j.1574-6968.2009.01489.x. [DOI] [PubMed] [Google Scholar]
- 35.Saiki K, Konishi K. 2010. The role of Sov protein in the secretion of gingipain protease virulence factors of Porphyromonas gingivalis. FEMS Microbiol Lett 302:166–174. 10.1111/j.1574-6968.2009.01848.x. [DOI] [PubMed] [Google Scholar]
- 36.Saiki K, Konishi K. 2010. Identification of a novel Porphyromonas gingivalis outer membrane protein, PG534, required for the production of active gingipains. FEMS Microbiol Lett 310:168–174. 10.1111/j.1574-6968.2010.02059.x. [DOI] [PubMed] [Google Scholar]
- 37.Chen YY, Peng B, Yang Q, Glew MD, Veith PD, Cross KJ, Goldie KN, Chen D, O'Brien-Simpson N, Dashper SG, Reynolds EC. 2011. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol Microbiol 79:1380–1401. 10.1111/j.1365-2958.2010.07530.x. [DOI] [PubMed] [Google Scholar]
- 38.Glew MD, Veith PD, Chen D, Gorasia DG, Peng B, Reynolds EC. 2017. PorV is an outer membrane shuttle protein for the type IX secretion system. Sci Rep 7:8790. 10.1038/s41598-017-09412-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauber F, Deme JC, Lea SM, Berks BC. 2018. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564:77–82. 10.1038/s41586-018-0693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heath JE, Seers CA, Veith PD, Butler CA, Nor Muhammad NA, Chen YY, Slakeski N, Peng B, Zhang L, Dashper SG, Cross KJ, Cleal SM, Moore C, Reynolds EC. 2016. PG1058 is a novel multidomain protein component of the bacterial type IX secretion system. PLoS One 11:e0164313. 10.1371/journal.pone.0164313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorasia DG, Veith PD, Hanssen EG, Glew MD, Sato K, Yukitake H, Nakayama K, Reynolds EC. 2016. Structural insights into the PorK and PorN components of the Porphyromonas gingivalis type IX secretion system. PLoS Pathog 12:e1005820. 10.1371/journal.ppat.1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naito M, Tominaga T, Shoji M, Nakayama K. 2019. PGN_0297 is an essential component of the type IX secretion system (T9SS) in Porphyromonas gingivalis: Tn-seq analysis for exhaustive identification of T9SS-related genes. Microbiol Immunol 63:11–20. 10.1111/1348-0421.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lasica AM, Goulas T, Mizgalska D, Zhou X, de Diego I, Ksiazek M, Madej M, Guo Y, Guevara T, Nowak M, Potempa B, Goel A, Sztukowska M, Prabhakar AT, Bzowska M, Widziolek M, Thogersen IB, Enghild JJ, Simonian M, Kulczyk AW, Nguyen KA, Potempa J, Gomis-Ruth FX. 2016. Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. Sci Rep 6:37708. 10.1038/srep37708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smalley JW, Olczak T. 2017. Heme acquisition mechanisms of Porphyromonas gingivalis - strategies used in a polymicrobial community in a heme-limited host environment. Mol Oral Microbiol 32:1–23. 10.1111/omi.12149. [DOI] [PubMed] [Google Scholar]
- 45.Berleman J, Auer M. 2013. The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environ Microbiol 15:347–354. 10.1111/1462-2920.12048. [DOI] [PubMed] [Google Scholar]
- 46.Kadurugamuwa JL, Beveridge TJ. 1996. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol 178:2767–2774. 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li ZS, Clarke AJ, Beveridge TJ. 1998. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol 180:5478–5483. 10.1128/JB.180.20.5478-5483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nudleman E, Wall D, Kaiser D. 2005. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 309:125–127. 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 49.Kadurugamuwa JL, Beveridge TJ. 1999. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other Gram-negative bacteria. Microbiology 145:2051–2060. 10.1099/13500872-145-8-2051. [DOI] [PubMed] [Google Scholar]
- 50.Vassallo C, Pathak DT, Cao P, Zuckerman DM, Hoiczyk E, Wall D. 2015. Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. Proc Natl Acad Sci U S A 112:E2939–E2946. 10.1073/pnas.1503553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 5:e1000382. 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gorasia DG, Chreifi G, Seers CA, Butler CA, Heath JE, Glew MD, McBride MJ, Subramanian P, Kjaer A, Jensen GJ, Veith PD, Re C. 2020. In situ structure and organisation of the type IX secretion system. bioRxiv 10.1101/2020.05.13.094771:2020.05.13.094771. [DOI]
- 53.Gabarrini G, Grasso S, van Winkelhoff AJ, van Dijl JM. 2020. Gingimaps: protein localization in the oral pathogen Porphyromonas gingivalis. Microbiol Mol Biol Rev 84:e00032-19. 10.1128/MMBR.00032-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gorasia DG, Glew MD, Veith PD, Reynolds EC. 2020. Quantitative proteomic analysis of the type IX secretion system mutants in Porphyromonas gingivalis. Mol Oral Microbiol 35:78–84. 10.1111/omi.12283. [DOI] [PubMed] [Google Scholar]
- 55.Bowden MG, Kaplan HB. 1998. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol Microbiol 30:275–284. 10.1046/j.1365-2958.1998.01060.x. [DOI] [PubMed] [Google Scholar]
- 56.Wall D, Kaiser D. 1999. Type IV pili and cell motility. Mol Microbiol 32:1–10. 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 57.Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat Rev Microbiol 2:946–953. 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 58.Weinberg ED. 2009. Iron availability and infection. Biochim Biophys Acta 1790:600–605. 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Kesavalu L, Holt SC, Ebersole JL. 2003. In vitro environmental regulation of Porphyromonas gingivalis growth and virulence. Oral Microbiol Immunol 18:226–233. 10.1034/j.1399-302X.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 60.Marsh PD, McDermid AS, McKee AS, Baskerville A. 1994. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalis W50. Microbiology 140:861–865. 10.1099/00221287-140-4-861. [DOI] [PubMed] [Google Scholar]
- 61.Gruss A, Borezee-Durant E, Lechardeur D. 2012. Environmental heme utilization by heme-auxotrophic bacteria. Adv Microb Physiol 61:69–124. 10.1016/B978-0-12-394423-8.00003-2. [DOI] [PubMed] [Google Scholar]
- 62.Bramanti TE, Holt SC. 1991. Roles of porphyrins and host iron transport proteins in regulation of growth of Porphyromonas gingivalis W50. J Bacteriol 173:7330–7339. 10.1128/jb.173.22.7330-7339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schifferle RE, Shostad SA, Bayers-Thering MT, Dyer DW, Neiders ME. 1996. Effect of protoporphyrin IX limitation on Porphyromonas gingivalis. J Endod 22:352–355. 10.1016/S0099-2399(96)80216-0. [DOI] [PubMed] [Google Scholar]
- 64.Shizukuishi S, Tazaki K, Inoshita E, Kataoka K, Hanioka T, Amano A. 1995. Effect of concentration of compounds containing iron on the growth of Porphyromonas gingivalis. FEMS Microbiol Lett 131:313–317. 10.1111/j.1574-6968.1995.tb07793.x. [DOI] [PubMed] [Google Scholar]
- 65.Olczak T, Simpson W, Liu XY, Genco CA. 2005. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol Rev 29:119–144. 10.1016/j.femsre.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 66.Lewis JP. 2010. Metal uptake in host-pathogen interactions: role of iron in Porphyromonas gingivalis interactions with host organisms. Periodontol 2000 52:94–116. 10.1111/j.1600-0757.2009.00329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tompkins GR, Wood DP, Birchmeier KR. 1997. Detection and comparison of specific hemin binding by Porphyromonas gingivalis and Prevotella intermedia. J Bacteriol 179:620–626. 10.1128/jb.179.3.620-626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL, Jr.. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 69.Zainal-Abidin Z, Veith PD, Dashper SG, Zhu Y, Catmull DV, Chen YY, Heryanto DC, Chen D, Pyke JS, Tan K, Mitchell HL, Reynolds EC. 2012. Differential proteomic analysis of a polymicrobial biofilm. J Proteome Res 11:4449–4464. 10.1021/pr300201c. [DOI] [PubMed] [Google Scholar]
- 70.Stacy A, Abraham N, Jorth P, Whiteley M. 2016. Microbial community composition impacts pathogen iron availability during polymicrobial infection. PLoS Pathog 12:e1006084. 10.1371/journal.ppat.1006084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10:497–506. 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bertani B, Ruiz N. 2018. Function and biogenesis of lipopolysaccharides. EcoSal Plus 8. 10.1128/ecosalplus.ESP-0001-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murray GL, Attridge SR, Morona R. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar typhimurium with macrophages and complement. J Bacteriol 188:2735–2739. 10.1128/JB.188.7.2735-2739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Man SM, Karki R, Kanneganti TD. 2017. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev 277:61–75. 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszyński A, Forsberg LS, Carlson RW, Dixit VM. 2013. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341:1246–1249. 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 77.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. 2014. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514:187–192. 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 78.Lagrange B, Benaoudia S, Wallet P, Magnotti F, Provost A, Michal F, Martin A, Di Lorenzo F, Py BF, Molinaro A, Henry T. 2018. Human caspase-4 detects tetra-acylated LPS and cytosolic Francisella and functions differently from murine caspase-11. Nat Commun 9:242. 10.1038/s41467-017-02682-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee K, Roberts JS, Choi CH, Atanasova KR, Yilmaz O. 2018. Porphyromonas gingivalis traffics into endoplasmic reticulum-rich-autophagosomes for successful survival in human gingival epithelial cells. Virulence 9:845–859. 10.1080/21505594.2018.1454171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Darveau RP, Pham TTT, Lemley K, Reife RA, Bainbridge BW, Coats SR, Howald WN, Way SS, Hajjar AM. 2004. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect Immun 72:5041–5051. 10.1128/IAI.72.9.5041-5051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coats SR, Pham TT, Bainbridge BW, Reife RA, Darveau RP. 2005. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J Immunol 175:4490–4498. 10.4049/jimmunol.175.7.4490. [DOI] [PubMed] [Google Scholar]
- 82.Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. 2009. Human Toll-like receptor 4 responses to P gingivalis are regulated by lipid A 1- and 4'-phosphatase activities. Cell Microbiol 11:1587–1599. 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Coats SR, Berezow AB, To TT, Jain S, Bainbridge BW, Banani KP, Darveau RP. 2011. The lipid A phosphate position determines differential host Toll-like receptor 4 pesponses to phylogenetically related symbiotic and pathogenic bacteria. Infect Immun 79:203–210. 10.1128/IAI.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hajishengallis G. 2014. The inflammophilic character of the periodontitis-associated microbiota. Mol Oral Microbiol 29:248–257. 10.1111/omi.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen YY, Cross KJ, Paolini RA, Fielding JE, Slakeski N, Reynolds EC. 2002. CPG70 is a novel basic metallocarboxypeptidase with C-terminal polycystic kidney disease domains from Porphyromonas gingivalis. J Biol Chem 277:23433–23440. 10.1074/jbc.M200811200. [DOI] [PubMed] [Google Scholar]
- 86.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526. 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 88.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res 28:235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eichinger A, Beisel HG, Jacob U, Huber R, Medrano FJ, Banbula A, Potempa J, Travis J, Bode W. 1999. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J 18:5453–5462. 10.1093/emboj/18.20.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Diego I, Ksiazek M, Mizgalska D, Koneru L, Golik P, Szmigielski B, Nowak M, Nowakowska Z, Potempa B, Houston JA, Enghild JJ, Thogersen IB, Gao JL, Kwan AH, Trewhella J, Dubin G, Gomis-Ruth FX, Nguyen KA, Potempa J. 2016. The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain. Sci Rep 6:23123. 10.1038/srep23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.