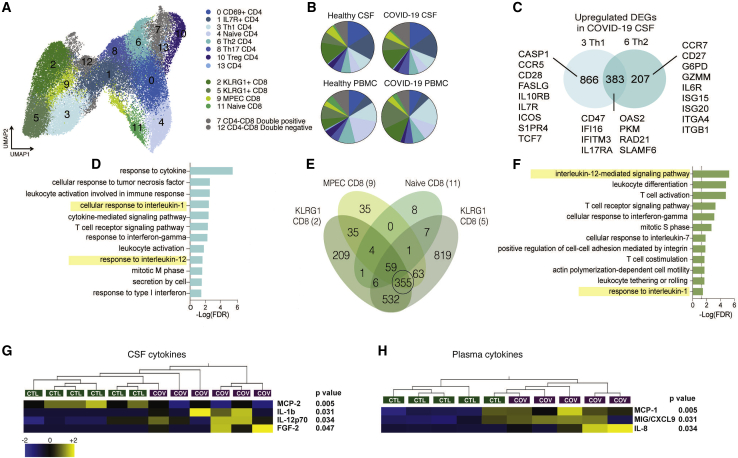
Figure 2.
Transcriptional characterization of T cells in CSF and PBMCs of individuals with COVID-19
(A) Reclustered UMAP projection of combined CSF and peripheral blood T cells, demonstrating CD4 and CD8 T cell subsets (two KLRG1+ clusters are distinguished by GZMB and IFNG expression; Figure S3).
(B) Pie charts depicting the relative population frequency of different T cell subtypes found in CSF and PBMCs of control individuals and those with COVID-19.
(C) Venn diagram depicting genes upregulated (adjusted p < 0.05) in CSF of individuals with COVID-19 compared with PBMCs of individuals with COVID-19 in Th1 and Th2 CD 4 T cells.
(D) Gene Ontology analysis of genes that are upregulated in Th1 and Th2 cells, as depicted in (C).
(E) Quad-Venn diagram of genes upregulated in CSF of individuals with COVID-19 compared with CSF of control individuals in CD8 T cells. Genes shared by the three effector CD8 T cell subtypes are circled.
(F) Gene Ontology analysis of genes shared between the three effector CD8 T cell subtypes in (F).
(G and H) Heatmap of Luminex-based cytokine profiling of CSF (G) and plasma (H) from individuals with COVID-19 and control individuals showing cytokines that were increased significantly increased in individuals with COVID-19 compared with control individuals (n = 6 CSF, n = 5 plasma). For each cytokine, two-tailed p values were calculated using Student’s t test. Data for each row were mean centered; each column shows data from one sample.