Figure 5 .
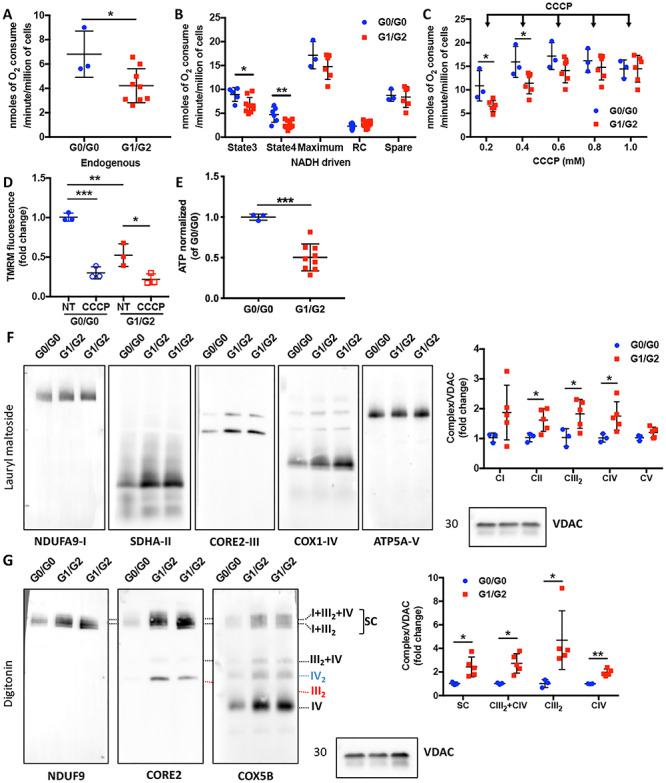
Podocytes carrying APOL1 G1/G2 risk alleles are characterized by mitochondrial dysfunction in association with increased OXPHOS complexes. (A and B) Scatter plot quantification of endogenous (A) and substrate-driven (B) oxygen consumption rates (OCR, nmol of oxygen consumed per minute per million of cells) in podocytes carrying G0/G0 and G1/G2 alleles (n = 3–9). (C) Scatter plot quantification of oxygen consumption rates during carbonyl cyanide m-chlorophenylhydrazone (CCCP) titrations. The arrows indicate sequential additions of CCCP (1 μl incremental addition of 0.1 M CCCP) (n = 3 & 6). (D) Scatter plot quantification of mitochondrial membrane potential in untreated (NT) and uncoupler agent (CCCP) treated podocytes carrying G0/G0 and G1/G2 (n = 3). (E) ATP content was determined in podocytes carrying G0/G0 and G1/G2 alleles using a luminescence assay and normalized to cell number (n = 3 & 9). (F) Representative Western blot images of BN-PAGE analysis using lysates obtained by lauryl maltoside extraction and probed for NDUFA9 (Complex I: CI), SDHA (CII), CORE2 (CIII), COX1 (CIV), ATP5A (CV) and VDAC was detected in SDS-PAGE as a loading control for the detection of steady state levels of individual complexes (left panel). Signals from the BN-PAGE analysis of lauryl maltoside extracts were quantified by densitometry and normalized to VDAC (right panel, n = 3 & 5). (G) Representative Western blot images of BN-PAGE analysis using lysates obtained by digitonin extraction and probed for CORE2, NDUFA9, COX5B and VDAC was detected in SDS-PAGE as a loading control to determine the OXPHOS complex distribution at the mitochondrial membrane (left panel). Signals from the BN-PAGE experiments of digitonin extracts were quantified by densitometry and normalized to VDAC (right panel, n = 3 & 5). The error bars represent mean ± SD of biologically independent experiments. Two-tailed Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
