Figure 2. Secondary Structural Model of Full Length 7SK snRNA as Determined by DMS-MaPseq.
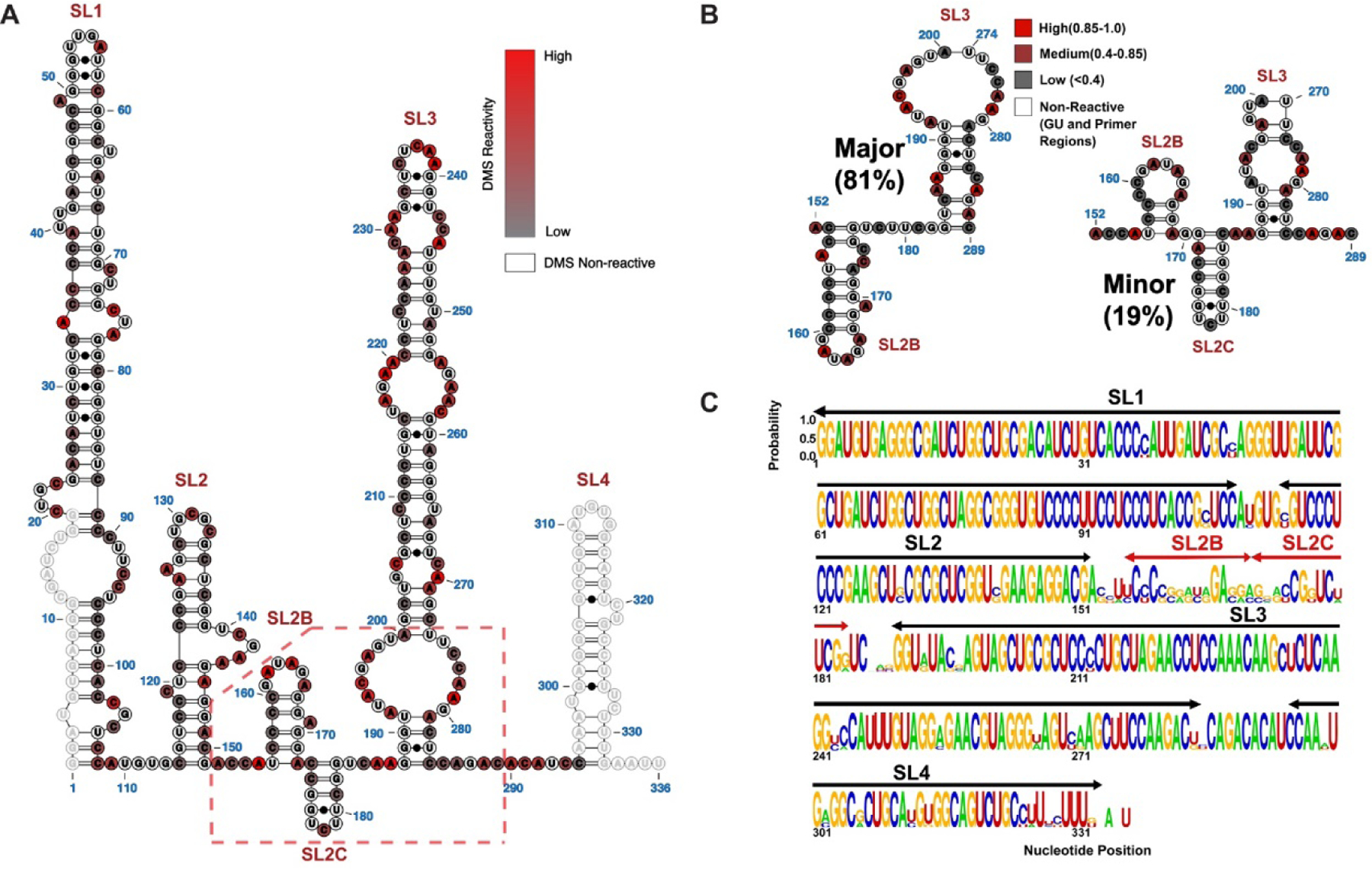
(A) The population averaged secondary structure of 7SK snRNA as determined by incorporating DMS reactivity indices as pseudoenergy restraints. The normalized indices are superimposed as a scaled color-code from 0 (low reactivity) to 1 (high reactivity) for each modified AC nucleotide. Primer regions and GU nucleotides are shown as open circles with no color code. (B) Two major 7SK snRNA conformers were derived from DREEM clustering of DMS-MaPseq reactivity indices. The color code is according to DMS reactivities of the nucleotides, coded as low (Grey), medium (Maroon) and high (Red). The significant structural differences localize to the SL2–3 linker and the base of SL3. (C) Logo plot analysis depicting the phylogenetic conservation of 7SK snRNA elements by aligning sequences from human, Mus musculus (M63671), Rattus norvegicus (K02909), chicken(AJ890101), zebra fish (AJ890102), Tetraodon nigrovidis (AJ890103) and Takifugu rubripes (AJ890104).
