Figure 6. A Conceptual Model to Interpret hnRNP A1/A2–7SK Assembly.
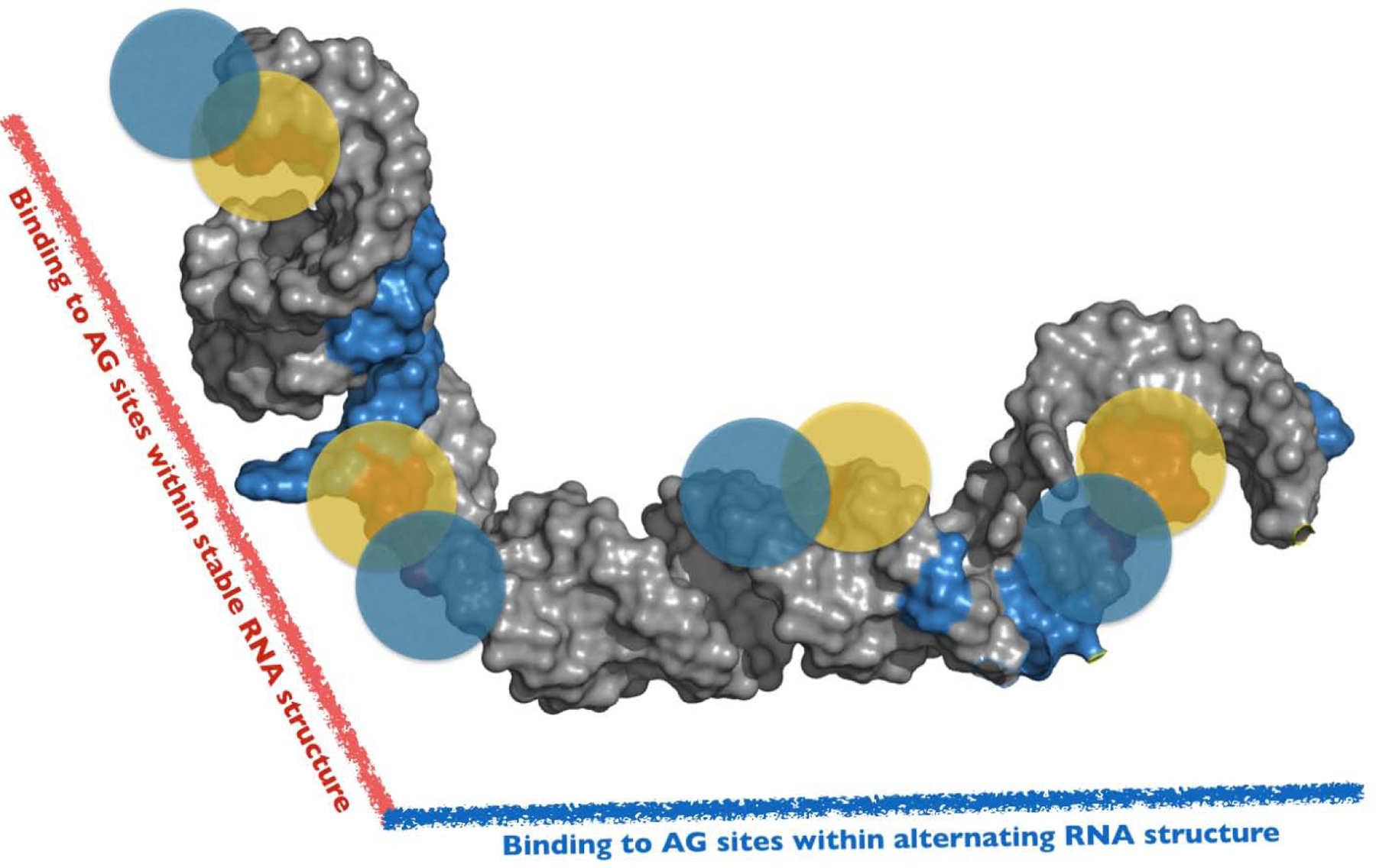
A 3D conceptual framework to interpret the complexities by which hnRNP A1/A2 proteins assemble along the SL3 domain of 7SK snRNA. The model is consistent with independent data sets derived from DMS-MaPseq, NMR, Calorimetry and SEC-SAXS measurements. The model posits that hnRNP A1/A2 proteins bind to specific 5’-Y/RAG-3’ sites positioned within distinct structural environments along the entire surface of SL3. Accordingly, the physicochemical properties of these local sites differentially modulate hnRNP A1/A2 interactions to form a functional 3D complex. The full-length SL3 (nts 188–184) structure shown here was determined using the FARFAR2 module of Rosetta 3.11 and (note this is not the experimentally-determined structural model of the shorter SL3M construct). Blue and Yellow circles represent the UP1 domain of hnRNP A1.
