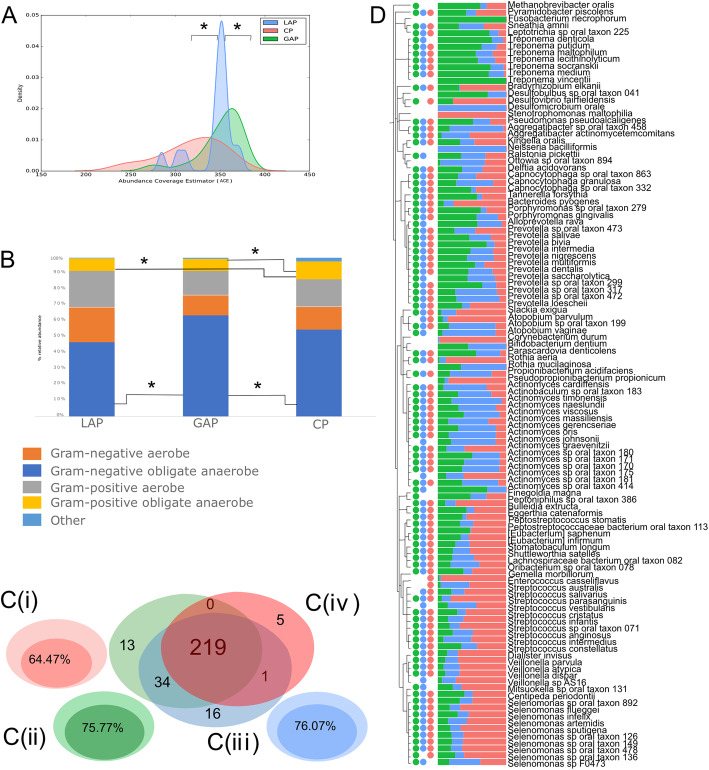
Fig. 3.
Disease-specific taxonomical indicators. Density curves of alpha diversity (ACE) are shown in a. The peak indicates the median values for each group, and the x-axis shows the data range. LAP exhibited significantly lower alpha diversity than the other 2 groups (p < 0.0001, Dunn’s test). Distribution of species-level taxa by gram staining characteristics and oxygen requirements in is shown in b. GAP patients demonstrated significantly greater gram-negative anaerobic bacteria and lower gram-negative aerobic bacteria when compared to the other two groups (p < 0.01, Dunn’s test for multiple comparisons). Percent of the microbiome that is shared by 80% or more of individuals (common core microbiome) with CP, GAP, and LAP are graphically indicated by the Euler graphs c (i–iii), as well as the number of core species shared by all three diseases is shown in c (iv). Phylogenetic tree of species that were significantly different between groups (p < 0.05, FDR-adjusted Wald test) are shown in 3D. Bars represent normalized mean relative abundances, while the solid circles indicate species that belong to the common disease core. Data supporting this figure can be found in Supplemental Table 1