Figure 1.
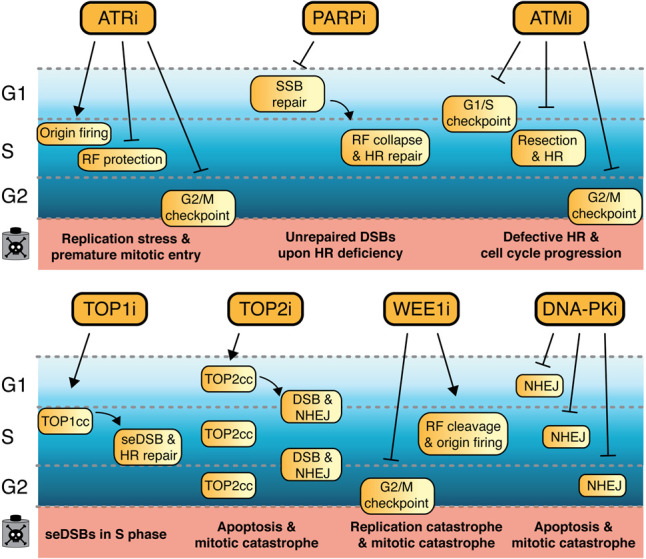
DDR inhibitors and their impacts on DSB repair and cell cycle progression. In mammalian cells, four main DSB pathways exist, which operate dependent on the stage of the cell cycle: nonhomologous end-joining (NHEJ), alternative end-joining (alt-EJ), homologous recombination (HR), and single-strand annealing (SSA) (Scully et al. 2019). TOP2i induced DSBs are predominantly repaired via NHEJ, which while being described as “error-prone,” ensures effective repair of broken DNA ends particularly during G0 and in the G1 phase of the cell cycle. DNA-PK is crucial for effective NHEJ, and DNA-PK inhibitors impair DSB repair via NHEJ. TOP1 inhibitors generate single-ended DSBs (seDSBs) in S phase, which require HR for accurate repair. HR is a form of DNA recombination where DNA homology and synthesis can accurately regenerate the sequence surrounding the DSB, facilitated by a sister chromatid as template and therefore restricted to S or G2 (Karanam et al. 2012). DNA damage arising from PARP inhibition also requires HR, since spontaneously occurring SSBs are converted into seDSBs during DNA replication upon PARP inhibitors (PARPis). In addition, PARPi cause mitotic catastrophe and induce DNA replication stress through altering DNA replication fork speed. A fundamental step during HR-mediated repair is DNA end resection, generating ssDNA overhangs that are rapidly covered by RPA and consequently by RAD51. ATR is activated by ssDNA as a result of DNA end resection or DNA polymerase uncoupling from helicase activity during DNA replication. Consequently, ATR inhibitors primarily have impacts during S and G2 phases of the cell cycle. Additionally, ATRi overrides the G2/M cell cycle checkpoint, therefore causing premature entry into mitosis. Importantly, ATM inhibition affects efficient HR, alongside its impact on G1/S and G2/M cell cycle checkpoints in response to DNA damage. Analogous to ATRi and ATMi, WEE1 inhibitors affect the G2/M cell cycle checkpoint. Moreover, WEE1i cause replication stress through dysregulated origin firing and cleavage of DNA replication forks, resulting in DSBs.
