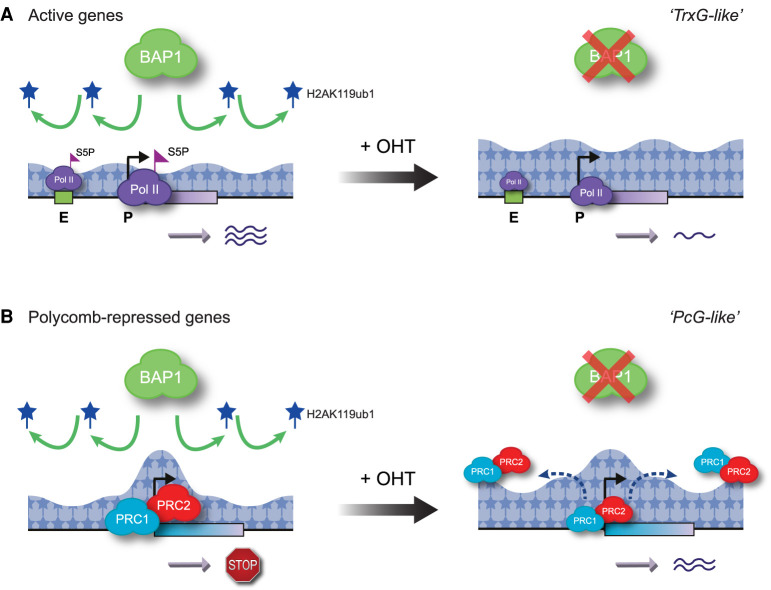
Figure 6.
A model illustrating how BAP1 can regulate gene expression by constraining pervasive H2AK119ub1. (A) BAP1 facilitates gene expression by constraining the pervasive sea of H2AK119ub1 that covers the genome. Inducible removal of BAP1 (+OHT) results in a broad accumulation of H2AK119ub1 throughout the genome. Elevated H2AK119ub1 indiscriminately counteracts Ser5 phosphorylation (S5P) on the CTD of Pol II at gene regulatory elements ([P] promoters, [E] enhancers), and causes widespread reductions in transcription and gene expression. This explains why disruption of BAP1 and other PR-DUB subunits can lead to Trithorax group (TrxG)-like phenotypes. (B) BAP1 also indirectly supports repression of a subset of Polycomb target genes by counteracting pervasive H2AK119ub1 and focusing high levels of Polycomb complexes at target gene promoters. In the absence of BAP1, PRC1/PRC2 occupancy at Polycomb target sites is reduced, presumably due to the increased binding of these complexes to elevated H2AK119ub1 elsewhere in the genome. This leads to derepression of a subset of Polycomb target genes that appear to rely on high-level Polycomb complex occupancy for their silencing, providing a potential molecular rationale for why the BAP1 ortholog in Drosophila has been originally characterized as a Polycomb group (PcG) gene.