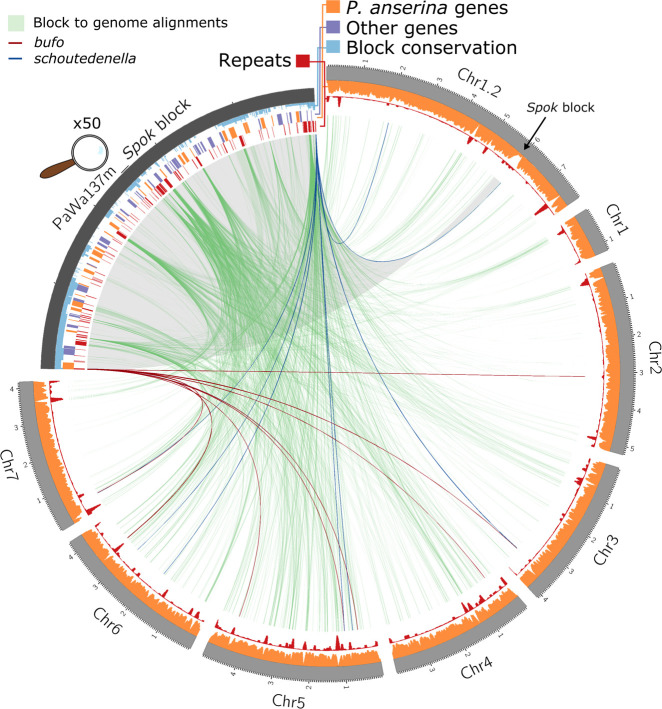
Figure 2.
A Circos plot of the genome of Wa137 (light gray track, size in Mb) aligned against its own Spok block (dark gray, enlarged by 50×). The tracks on the Spok block, from outside inward, represent: the conservation of regions among the different iterations of the Spok block where the height of the conservation track is equivalent to the number of blocks that have a given position (light blue); gene models from manual annotation of the Spok block, indicating genes with homologs within the reference genome of P. anserina (orange) and those without homologs (purple); and annotated repeated elements (red). The tracks on the chromosomal scaffolds show the coverage of genic regions (orange) and repeats (red) calculated in sliding windows of 50 kb with steps of 10 kb. Green lines connect homologous segments based on MUMmer alignments. The unclassified repetitive elements bufo and schoutedenella are connected in dark red and blue segments, respectively, based on BLASTN searches, highlighting potential past Spok block or closely related Enterprise element insertion points.