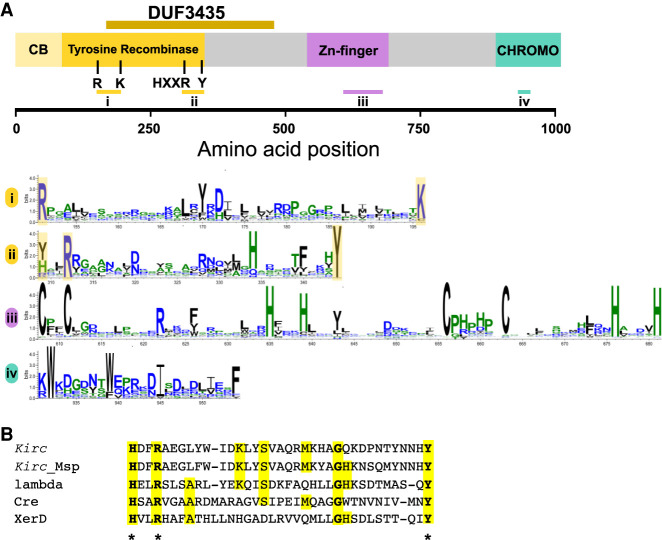
Figure 4.
Molecular characterization of Kirc. (A) Domain annotation of the Kirc protein obtained using HHPred. Four domains were identified: an N-terminal tyrosine recombinase domain with the associated core-binding domain, a central Zn-finger domain, and a C-terminal chromodomain. An alignment of 279 Kirc homologs generated by Gremlin with HHBlits was used to create sequence logo of relevant regions of the Kirc sequences, including the active site regions of the tyrosine recombinase domain (i and ii), the Zn-finger region (iii), and the chromodomain region (iv) using WebLogo. Residues corresponding to the catalytic pentad of the tyrosine recombinase are highlighted in yellow. (B) Alignment of the Kirc sequence with an active site region of known tyrosine recombinases together with the sequence of a Kirc homolog from Melanconium sp. strain NRRL 54901. Alignment is based on the HHPred output. Active site residues are marked with an asterisk. (Kirc) Kirc gene in the Spok block of strain Wa58 (sites 309–344), (Kirc_Msp) CE55503_10527 (110–145), (lambda) lambda integrase PDB: 5J0N (308–342), (Cre) recombinase enterobacteriaphage P1, PDB: 1XO0 (270–305), (XerD) XerD site-specific recombinase E. coli, PDB: 1A0P (242–277).