Fig. 1 |. Localization and deposition pathways of histone variants across the genome.
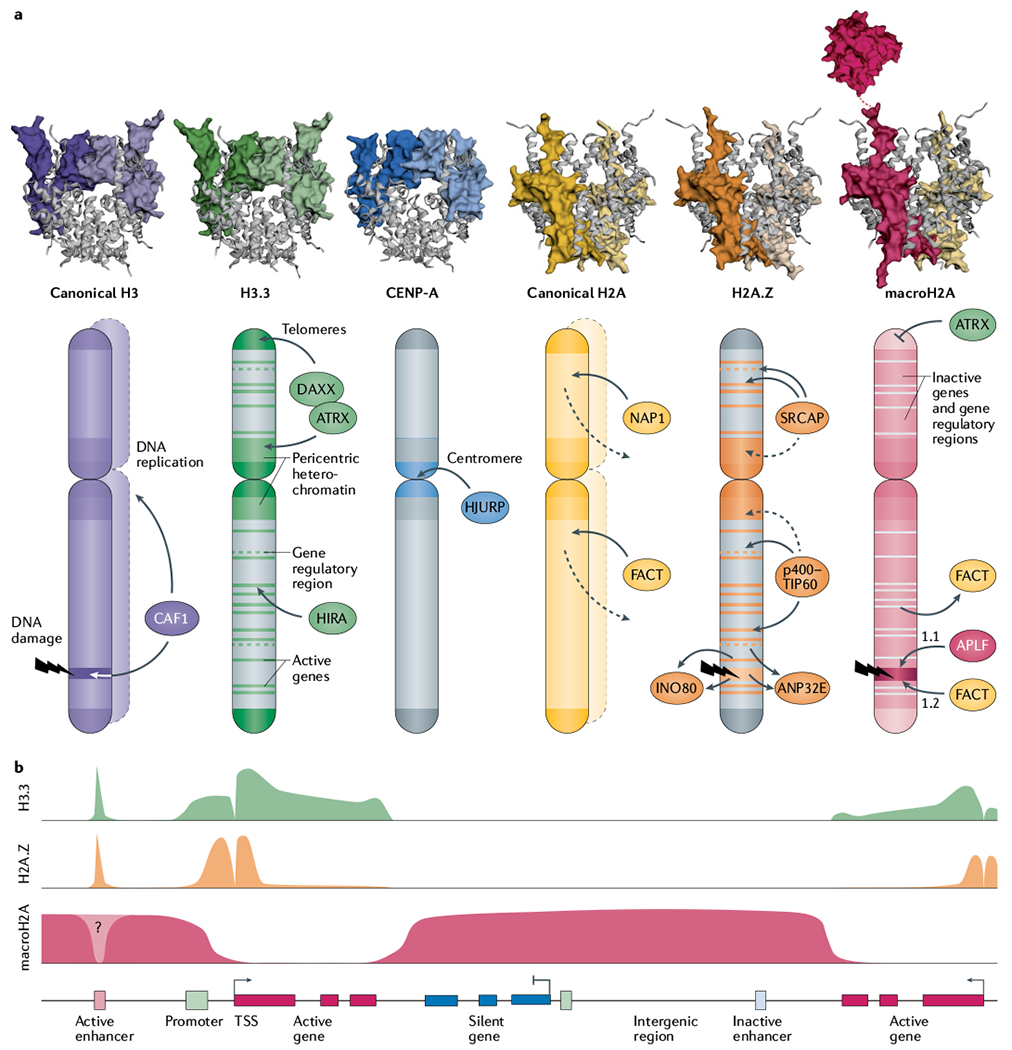
a | Histone variants are structurally similar to their canonical counterparts, with the exception of macroH2A’s non-histone macrodomain, which protrudes from the nucleosome particle via its linker region. Histone variants are differentially enriched across genomic landmarks as a function of histone chaperones. The CAF1 (chromatin assembly factor 1) complex deposits canonical H3 variants H3.1 and H3.2 during DNA replication or repair. H3.3 is deposited at active genes and at gene regulatory and nucleosome-depleted regions by the HIRA (histone cell cycle regulation-defective homologue A) complex, and at pericentric heterochromatin and subtelomeric regions by death domain-associated protein 6 (DAXX)–ATRX (α-thalassaemia/mental retardation syndrome X-linked). Centromeric protein A (CENP-A) is deposited at the active centromere by Holliday junction recognition protein (HJURP). NAP1 and the facilitates chromatin transcription (FACT) complex catalyse incorporation of canonical H2A during replication, and throughout the cell cycle, counterbalancing its continuous turnover (dashed lines). H2A.Z is deposited at active genes and regulatory regions by the SNF2-related CBP activator protein (SRCAP) and p400–TIP60 complexes, but is also enriched at pericentric heterochromatin through an unclear mechanism. The INO80 remodeling complex and ANP32E chaperone exchange H2A.Z for H2A at active genes, gene regulatory elements and DNA damage sites. APLF and FACT promote the accumulation of macroH2A1.1 and macroH2A1.2, respectively, at DNA damage sites. MacroH2A enrichment is negatively regulated by ATRX at telomeres and by FACT-mediated active eviction at transcribed genes. Crystal structures of canonical histones and H3.3 (REF.283), CENP-A284, H2A.Z12, macroH2A–H2A heterotypic nucleosome285 and macroH2A macrodomain76 were rendered with use of EzMol286. b | At gene-level resolution, H3.3 and H2A.Z are enriched at active regulatory elements (enhancers) and around the promoter of active genes, with the exception of the nucleosome-free region at the transcription start site (TSS). H3.3 is additionally present across transcribed gene bodies and transcription stop sites. MacroH2A forms large domains limited by actively transcribed regions.
