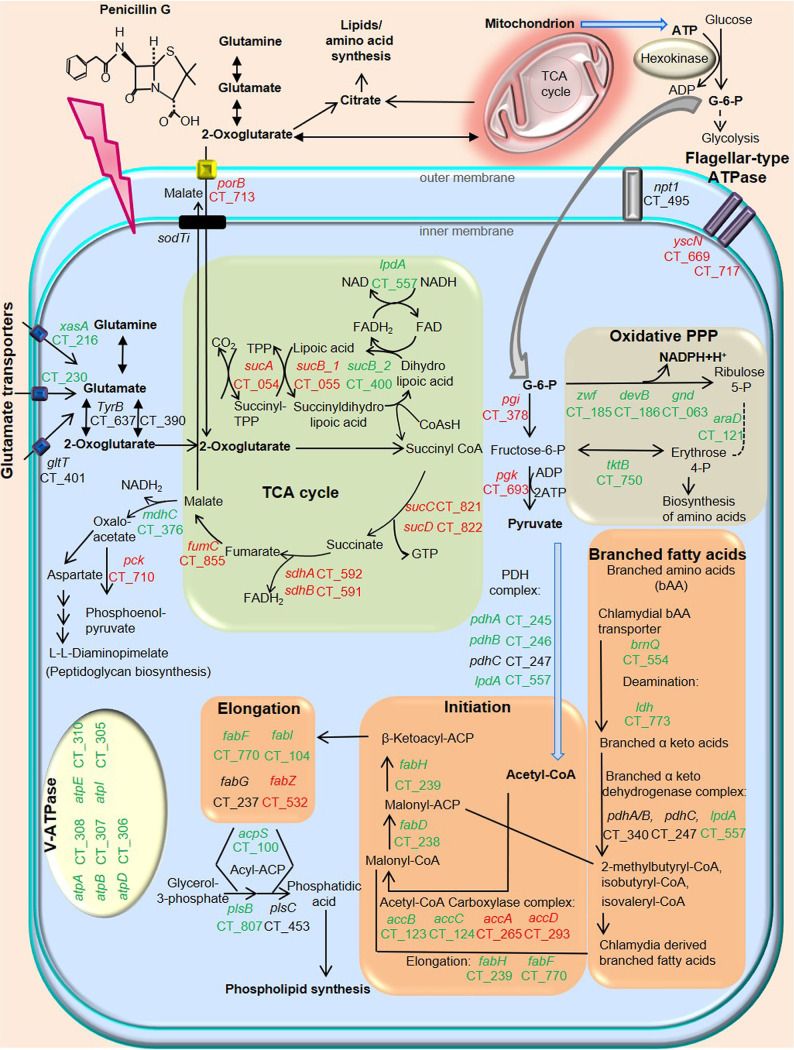
FIG 3.
Schematic representation of the metabolic pathways of C. trachomatis during treatment with penicillin. Genes marked in green were significantly upregulated, while genes marked in red were downregulated, in penicillin-induced chlamydial persistence compared to productive infection (Padj ≤ 0.05). Chlamydial energy production-associated genes are downregulated in penicillin-induced chlamydial persistence. The chlamydial substrate-specific porin PorB (porB), TCA cycle-related genes, and flagellar-type ATPase are downregulated in penicillin-induced chlamydial persistence. Hexokinase of mammalian cells requires mitochondrial ATP to catalyze the conversion of glucose into G-6-P, an essential substrate for chlamydial glycolysis. While chlamydial G-6-P isomerase (pgi) is downregulated, the rate-limiting enzyme G-6-P dehydrogenase (zwf) in the chlamydial PPP is upregulated in penicillin-induced chlamydial persistence, indicating that G-6-P could be mainly used for the chlamydial PPP. Furthermore, chlamydial fatty acid/phospholipid synthesis as well as acetyl-CoA synthesis are upregulated in penicillin-induced chlamydial persistence. This upregulation is linked to chlamydial phospholipid synthesis. Both acetyl-CoA and oxidative PPP-derived specific NADPH are essential for fatty acid synthesis. FAD, flavin adenine dinucleotide; FADH2, reduced flavin adenine dinucleotide; ACP, acyl carrier protein; CoA, coenzyme A.