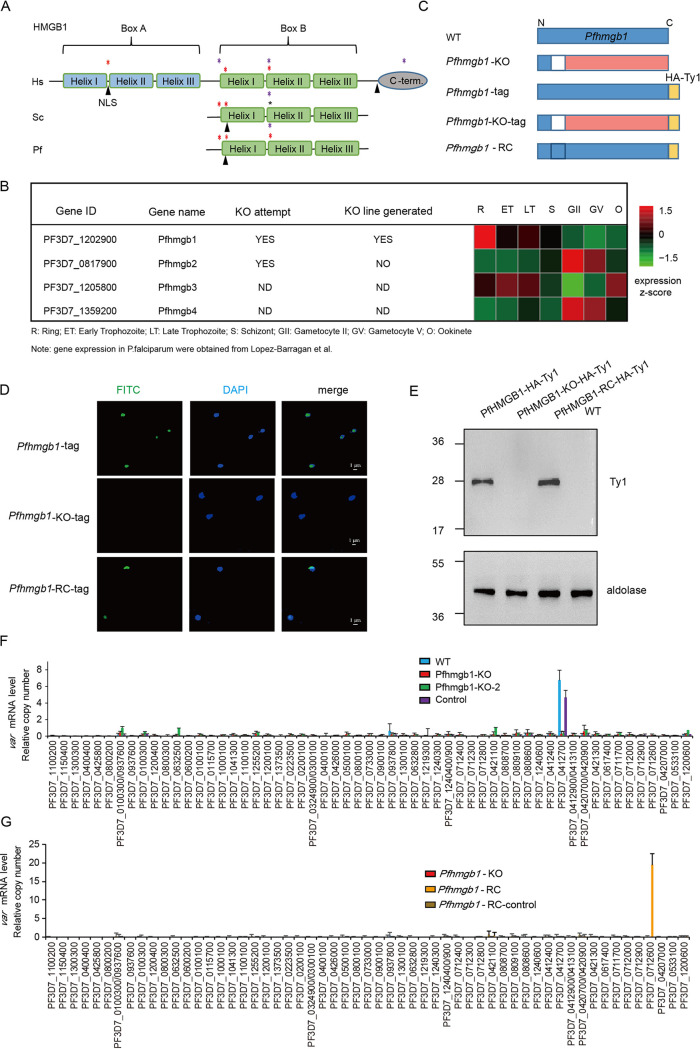
FIG 1.
Pfhmgb1-knockout dysregulates variant gene expression. (A) Schematic representation of HMGB1 proteins of Homo sapiens (Hs), Saccharomyces cerevisiae (Sc), and P. falciparum (Pf). The two boxes and individual α-helix domains are shown in parallel. The purple asterisks indicate the determinants for structure-specific recognition, and red asterisks indicate critical intercalating sites. The sites of putative nuclear localization signals (NLS) are indicated with each protein. (B) Transcriptional profiles and CRISPR-Cas9 knockout attempts of the four Pfhmgb genes in the 3D7 strain. Transcriptional abundance data were obtained from Lopez-Barragan et al. (51). (C) Schematic representation of generation of Pfhmgb1-KO, Pfhmgb1-HA-Ty1, Pfhmgb1-KO-HA-Ty1, and Pfhmgb1-RC transgenic parasites by CRISPR-Cas9. The blank region represents the deleted fragment and the pink region represents the disrupted ORF of the Pfhmgb1 gene. (D) Subcellular location of PfHMGB1 in transgenic ring-stage parasites detected by IFA with anti-Ty1 antibody (green). The nuclei are stained by 4′,6-diamidino-2-phenylindole (DAPI). (E) Western blot assay of PfHMGB1 protein in different clones. (F) RT-qPCR result for var genes in Pfhmgb1-KO (two clones from independent transfections), vector control, and WT clones. Error bars represent the standard errors of the means (SEMs) from two biological replicates. (G) RT-qPCR result for var genes in Pfhmgb1-KO, Pfhmgb1-RC, and transfection control (vector) clones. Error bars represent the SEMs for two biological replicates.