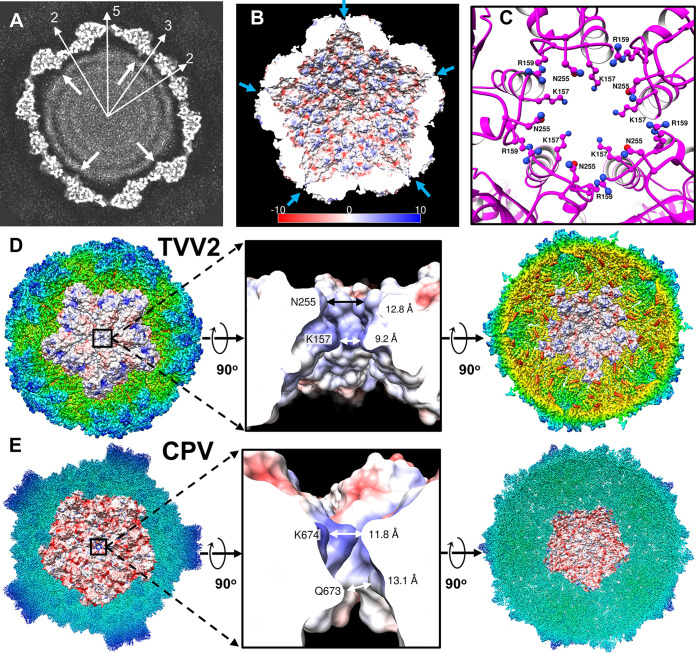
FIG 4.
TVV2 capsid coordinates genome arrangement and egression of transcription intermediates. (A) 10 Å thick slab view of the Icosahedral viral capsid of arbitrary contour colored in grayscale with map densities in white and 0-values in black. Sites of icosahedral symmetry through the slab are labeled with thin, white arrows, and thick, white arrows indicate the location of the C terminus based on the atomic model. Distance between dsRNA strands measures ∼30 Å. (B) Cutaway view of capsid interior with electrostatic potential diagrams illustrating positive (blue) and negative (red) surfaces. I5 vertices are marked with arrows (blue). (C) I5 vertex as viewed from the capsid exterior; CSP-A K157, R159, and N255 residues are shown as ball and stick diagrams. (D) Exterior view of the TVV2 capsid protein decamer fit into the icosahedral map (radially colored) with surface colored by electrostatic potential (left). 90° rotation of A and cutaway view of the 5-fold channel, with local residues from a CSP-A subunit (K157, R159, and N255) and channel diameters labeled (middle). Interior view of TVV2 decamer surface fit into map (right). (E) Exterior view of the dsRNA virus CPV CSP in the quiescent state (RCSB: 3JAZ) decamer fit into the icosahedral map with surface colored based on electrostatic potential (left). A cutaway view of the 5-fold channel, with involved residues from an CSP-A conformer (K674 and Q673) and channel diameters labeled (middle). Interior view of CPV CSP decamer fit into map (right).