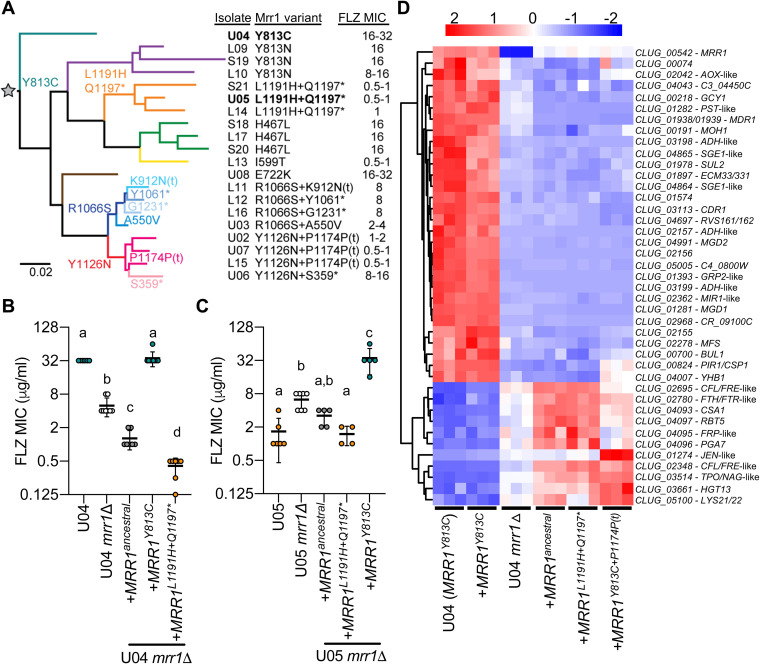
FIG 1.
Constitutively active and low-activity Mrr1 variants naturally evolved in a single C. lusitaniae population. (A) Maximum likelihood-based phylogeny constructed from SNPs identified in the whole-genome sequences of 20 C. lusitaniae isolates; modified from Demers et al. (10). Select branchpoints are marked with the Mrr1 variants (text colored to match branches) present in subsequent isolates. Mrr1 variants are identified by amino acid changes that resulted from SNPs or indels; * indicates a stop codon. The one-nucleotide indel in codons P1174 (insertion) and K912 (deletion) cause frameshift mutations that resulted in early termination, denoted with “(t),” at N1176 and L927, respectively. Gray star at the root of the tree denotes the “ancestral” MRR1 sequence, which lacks any of the mutations listed. U04 and U05, which are used in panels B and C, are highlighted. FLZ MICs (μg/ml) as determined in reference 10 are listed. (B) FLZ MICs for unaltered, mrr1Δ and MRR1 complemented strains in the FLZ-resistant U04 (native allele MRR1Y813C) strain background. (C) Same as in panel B, but in the FLZ-sensitive U05 strain background (native allele MRR1L1191H+Q1197*). Strains containing the same MRR1 allele in panels B and C are represented by circles of the same color. Data shown represent at least four independent assays on different days. Each sample was statistically compared to every other sample; the same lowercase letters indicate samples that are not significantly different, and different lowercase letters indicate significant differences (P < 0.0001 [B] or P < 0.001 [C]) as determined by one-way ANOVA with Tukey’s multiple-comparison test of log2-transformed values. (D) Differentially expressed genes between strains harboring the constitutively active Mrr1-Y813C variant (U04 and U04 mrr1Δ+MRR1Y813C) and those lacking MRR1 (U04 mrr1Δ) or harboring low-activity variants (U04 mrr1Δ+MRR1ancestral and U04 mrr1Δ+MRR1L1191H+Q1197*) when grown in liquid YPD medium; statistical cutoffs used were FDR of <0.05 and fold change of ≥2 (see Table S1 in the supplemental material). Normalized counts per million (CPM) from RNA-Seq are scaled by row (gene) with hierarchical clustering by Euclidean distance. Complemented strains are denoted by their respective MRR1 allele. Predicted C. albicans homologs are listed next to C. lusitaniae gene names (Table S1).