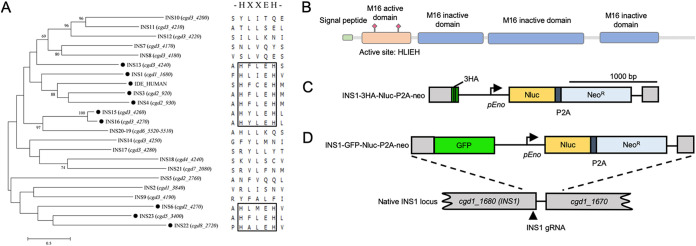
FIG 1.
Insulinase-like proteases in Cryptosporidium parvum and construction of C. parvum INS1-3HA and INS1-GFP strains. (A) Phylogenetic relationship of C. parvum insulinase-like protease family and human insulinase. The tree was constructed by a maximum likelihood analysis with 1,000 replications for bootstrapping. The active site HXXEH in each INS was defined by multiple alignments. The black dot indicates proteins with a predicted active site. Scale, 50 changes per 100 residues. (B) Domain architecture of INS1 showing the presence of one M16 active domain containing the active site HLIEH and three inactive domains. (C) Diagram of INS1-3HA tagging strategy. Construct was designed to add a 3HA tag and Nluc-P2A-NeoR cassette at the C terminus of INS1 (cgd1_1680). P2A, split peptide. INS1 gRNA marks the site of guide RNA homology. (D) Diagram of INS1-GFP tagging strategy. Construct was designed to add a GFP tag and Nluc-P2A-NeoR cassette at the C terminus of INS1 (cgd1_1680). P2A, split peptide; INS1 gRNA, site of guide RNA homology.