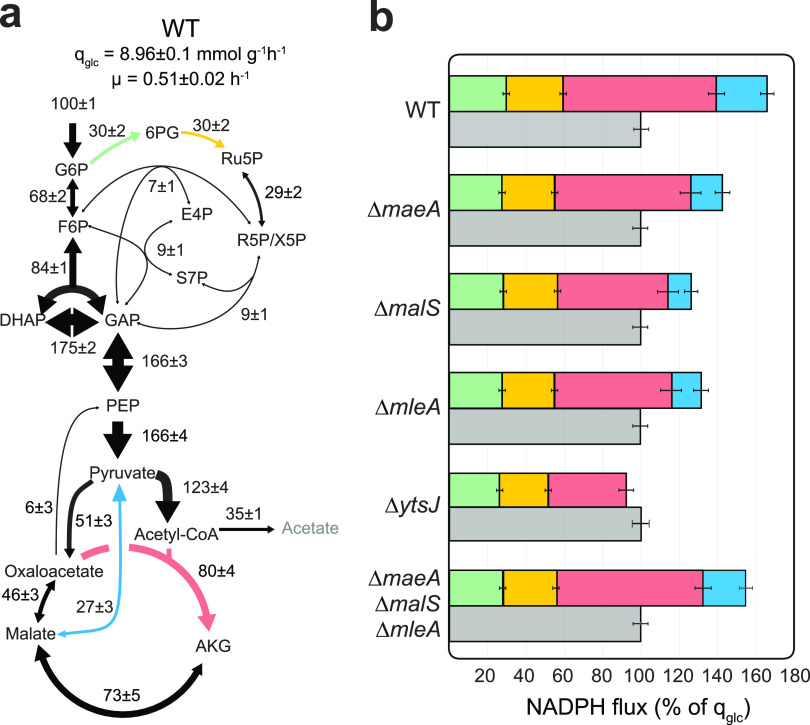
FIG 1.
Cellular NADPH balance. (a) Metabolic flux distribution in B. subtilis wild type, estimated by 13C flux ratio analysis and metabolite balancing. Flux values are normalized to the glucose uptake rate (qglc; mmol gCDW−1 h−1), and arrow sizes scale with flux magnitudes. (b) NADPH balances of wild-type and malic enzyme deletion strains. NADPH production is calculated by summing the flux rates of NADPH-dependent glucose-6-phosphate dehydrogenase (green), 6-phosphogluconate dehydrogenase (yellow), isocitrate dehydrogenase (red), and NADP+ specific malic enzyme (blue) and compared to normalized growth-dependent NADPH consumption (gray).