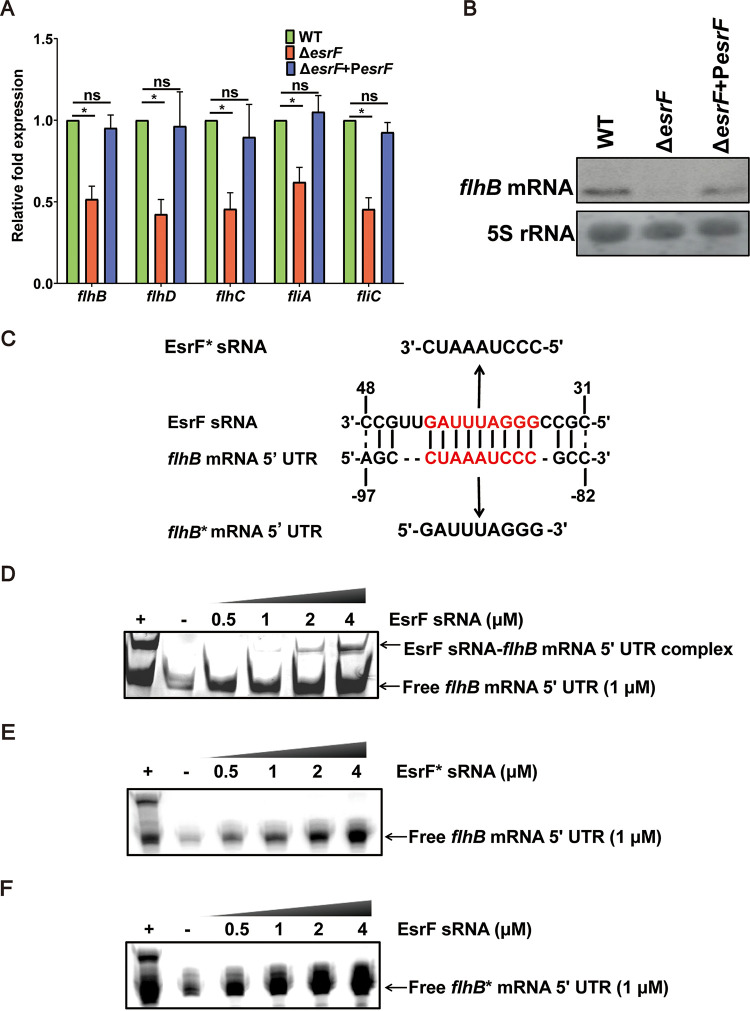
FIG 3.
Positive regulation of the downstream gene, flhB, by EsrF binding. (A) qRT-PCR of the expression of flhB, flhD, flhC, fliA, and fliC in WT, ΔesrF, and ΔesrF+PesrF strains. (B) Northern blotting was performed with a specific probe directed against the flhB mRNA in O157 WT, ΔesrF, and ΔesrF+PesrF strains; 5S rRNA was used as the loading control. (C) Predicted EsrF sRNA-flhB mRNA 5′ UTR base pairing. The lines and dashed lines indicate the predicted base pairs and incomplete base pairs between EsrF and flhB RNA 5′ UTR, respectively. The red letters represent the predicted, binding motif for EsrF action. EsrF* sRNA represents EsrF with point mutation with the GGGAUUUAG motif mutated to CCCUAAAUC, while flhB* mRNA 5′ UTR represents flhB with point mutations for which the motif CUAAAUCCC was mutated to GAUUUAGGG. (D) RNA-RNA EMSA of EsrF sRNA and flhB mRNA 5′ UTR. The EsrF complement strand (+) and yeast RNA (−) were used as positive and negative controls, respectively. (E) RNA-RNA EMSA of EsrF* sRNA and flhB mRNA 5′ UTR. (F) RNA-RNA EMSA of EsrF sRNA and flhB* mRNA 5′ UTRs. The data are presented as means ± the SD (n = 3; ns, no significance; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). All P values were calculated using a Student t test.