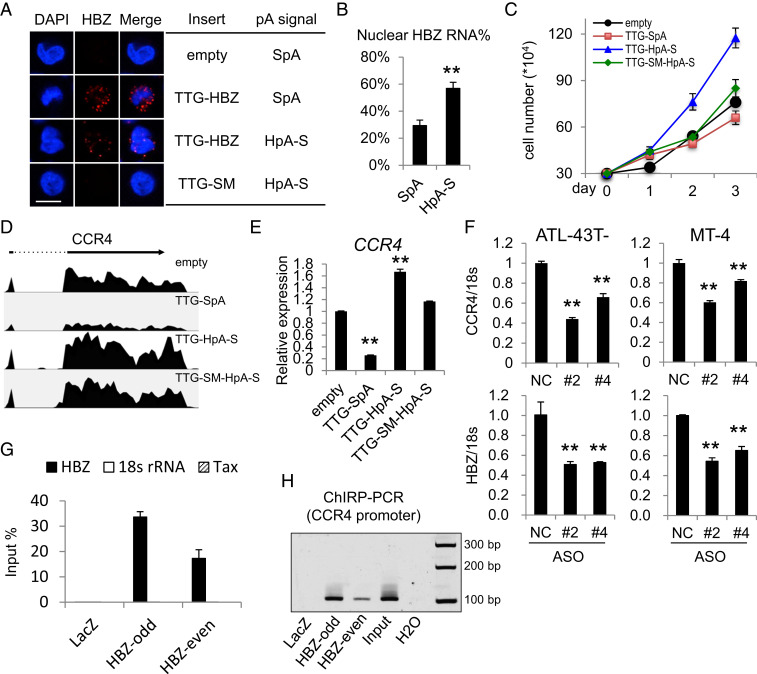
Fig. 3.
Nuclear-localized HBZ RNA associates with the CCR4 promoter and enhances CCR4 transcription. (A) Representative RNA-FISH results of four Kit225 lines. HBZ RNA was stained in red (scale bar, 10 μm). (B) The percentage of HBZ RNA that is nuclear in the TTG-SpA (SpA) and TTG-HpA-S (HpA-S) Kit225 lines. (C) A total of 3 × 104 cells of each line were seeded in full RPMI media with 100 U/mL IL-2, and cell number was counted every day until day three. (D) CCR4 tracks from RNA-seq results of the four cell lines. (E) qPCR verification of the RNA-seq results in D. (F) CCR4 and HBZ RNA levels in ASO-mediated HBZ-knockdown cells (#2 and #4) and negative control cells (NC). (G) qPCR result showing the retrieval efficiency of two pools of HBZ RNA probes (HBZ-odd and HBZ-even) and the negative control LacZ probes used in the ChIRP assay. Nonspecific retrieval of Tax RNA or 18s ribosomal RNA was not observed. (H) ChIRP-PCR was performed to amplify the previously reported CCR4 promoter region (−326 ∼ −220 bp) (17). Note that the negative control LacZ probes yielded no band whereas both pools of HBZ RNA probes yielded bands with the same size as input. The statistical analyses were performed by Student’s t test. **P < 0.01.