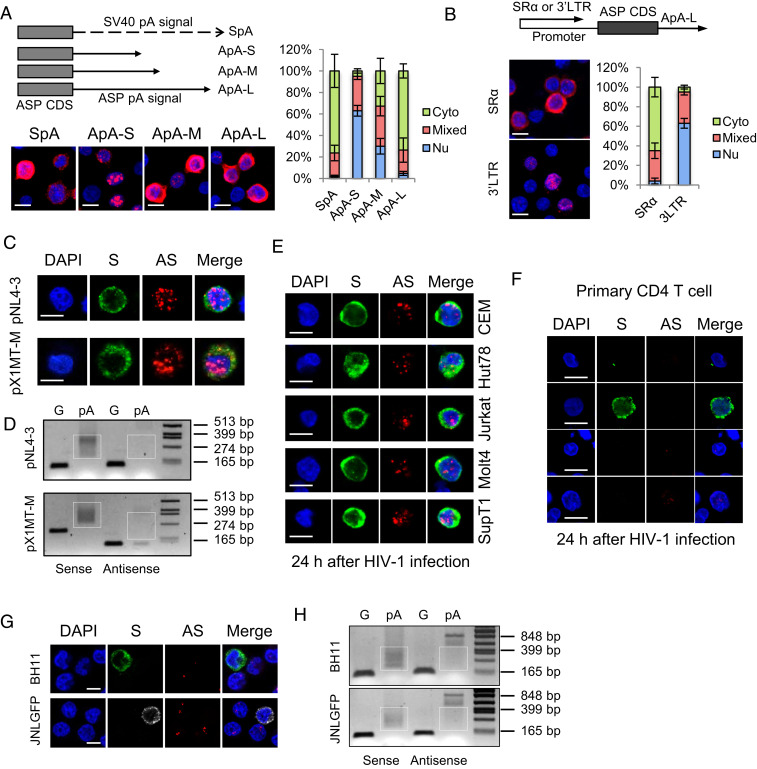
Fig. 4.
HIV-1 antisense mRNA, ASP, is inefficiently polyadenylated and predominantly localized in the nucleus in infected cells. HeLa cells were transfected with ASP-expression vectors, HIV-1–infectious clone pNL4-3 or HTLV-1–infectious clone pX1MT-M, and RNA-FISH (A–C) and a pA tail assay (D) were performed. (A) The Left Top illustrates ASP-expression vectors with modified polyadenylation signals from the ASP gene. ApA is cloned from the native ASP pA signal. ApA-S ends at the cleavage site of ApA, ApA-M ends at the G/T-rich region, and ApA-L ends 100 bp downstream the AATAAA sequence of ApA. See SI Appendix, Fig. S7A for more details. The Left Bottom shows representative RNA-FISH results from the transfected HeLa cells detecting ASP RNA (red). The percentage of cells with predominantly cytoplasmic, nuclear, or mixed localization patterns of ASP RNA was counted using ImageJ and is shown on the Right (scale bar, 10 μm). (B) RNA-FISH suggests the use of HIV-1 3′ LTR as the promoter caused nuclear retention of ASP RNA. Bar = 10 μm. (C) Representative RNA-FISH results from the transfected HeLa cells detecting viral sense (green) and antisense (red) mRNAs (scale bar, 10 μm). (D) pA tail assay results of transfected HeLa indicating efficient polyadenylation of viral sense mRNAs but inefficient polyadenylation of antisense mRNAs. Smears indicating pA products are marked by white boxes. The assay was preoptimized in order to obtain equivalent amounts of gene-specific products for sense and antisense mRNAs (SI Appendix, Fig. S7B). (E) Representative RNA-FISH results of five T cell lines de novo infected with HIV-1. HIV-1 sense mRNAs are shown in green, and ASP RNA is shown in red (scale bar, 10 μm). (F) Representative RNA-FISH results of primary human CD4 T cells de novo infected with HIV-1. HIV-1 sense mRNAs are shown in green, and ASP RNA is shown in red (scale bar, 10 μm). (G) Representative RNA-FISH results of BH11 or JNLGFP cells. HIV-1 sense mRNAs are shown in green, except in JNLGFP they are stained in white to avoid interference from GFP. HIV-1 antisense mRNA is shown in red (scale bar, 10 μm). (H) pA tail assay results of BH11 and JNLGFP cells indicating efficient polyadenylation of HIV-1 sense mRNAs but inefficient polyadenylation of antisense mRNAs. Smears indicating pA products are marked by white boxes. The assay was preoptimized in order to obtain equivalent amounts of gene-specific products for sense and antisense mRNAs (SI Appendix, Fig. S7D).