Figure 2. Cryo-EM structures of class 2 C002 and C121 hNAbs show binding to “up” and “down” RBDs.
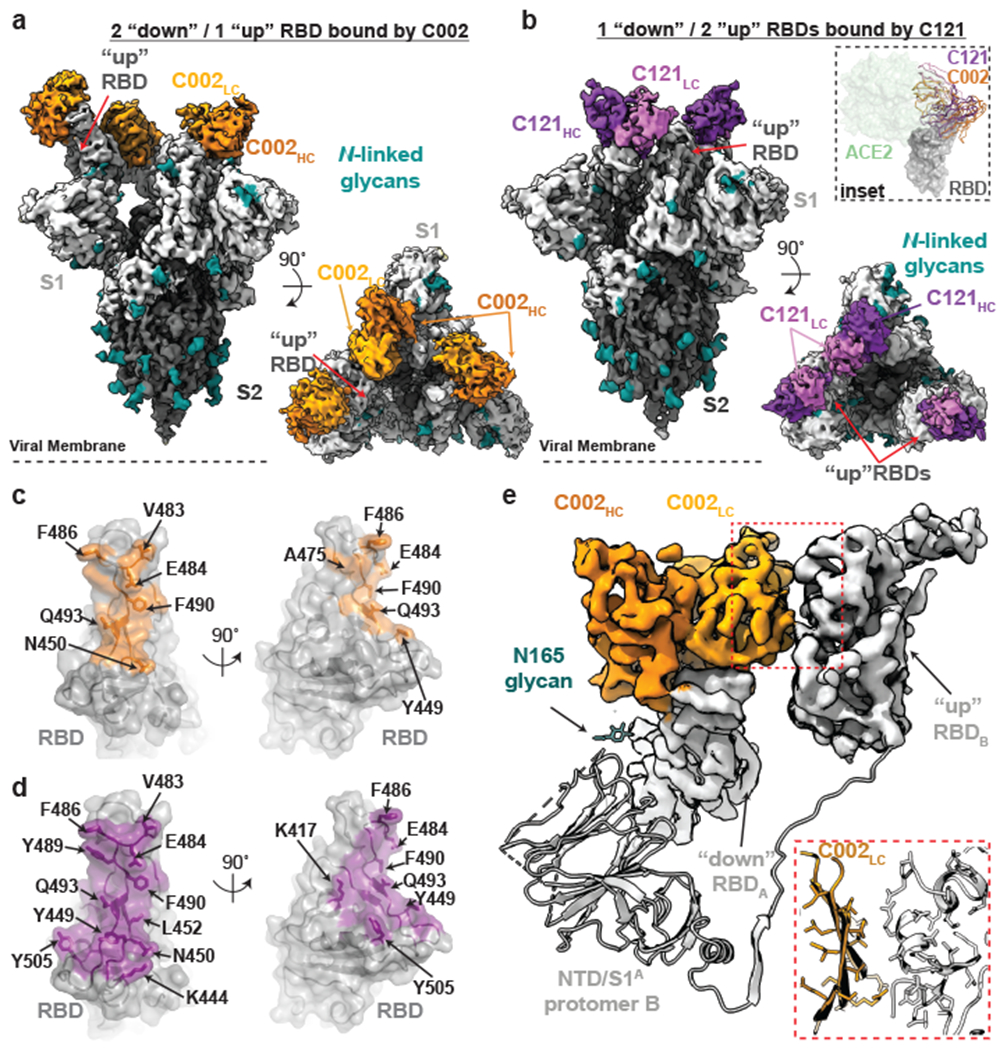
a,b, Cryo-EM densities for C002-S (panel a; 3.4Å) and C121-S complexes (panel b; 3.7Å) revealing binding of C002 or C121 to both “down” and “up” RBDs. Inset: Alignment of C002 and C121 Fabs on the same RBD. ACE2 is represented as a green surface for reference. c,d, Surface representations of C002 epitope (orange, panel c) and C121 epitope (purple, panel d) on the RBD surface (gray). RBD epitope residues (defined as residues containing atom(s) within 4Å of a Fab atom) are labeled in black. e, C002 forms inter-protomer contacts via binding to an adjacent “up” RBD conformation on the surface of the trimer spike (also observed for class 2 C121-, C119-, and C104-S structures, see Extended Data Fig.5). Red box: Close-up of adjacent “up” RBD and C002 LC interface.
