Infections caused by nontuberculous mycobacteria (NTM) are increasing globally. Mycobacterium avium complex (MAC) and Mycobacterium abscessus complex are the most frequently encountered NTM, and oral treatment options are extremely limited for these pathogens, especially for the M. abscessus complex. In this study, the in vitro potency of omadacycline, a new tetracycline derivative, was tested against 111 isolates of NTM.
KEYWORDS: nontuberculous mycobacteria, omadacycline, susceptibility testing
ABSTRACT
Infections caused by nontuberculous mycobacteria (NTM) are increasing globally. Mycobacterium avium complex (MAC) and Mycobacterium abscessus complex are the most frequently encountered NTM, and oral treatment options are extremely limited for these pathogens, especially for the M. abscessus complex. In this study, the in vitro potency of omadacycline, a new tetracycline derivative, was tested against 111 isolates of NTM. MIC testing was performed as recommended by the Clinical and Laboratory Standards Institute against 70 isolates of rapidly growing mycobacteria (RGM), of which >90% were tetracycline resistant. These included M. abscessus subsp. abscessus (20 isolates), M. abscessus subsp. massiliense (3), Mycobacterium chelonae (15 isolates), Mycobacterium immunogenum (7 isolates), the Mycobacterium fortuitum group, including six doxycycline-resistant isolates (12 isolates), and the Mycobacterium mucogenicum group, including four doxycycline-resistant isolates (10 isolates). Forty-one isolates of slowly growing mycobacteria (SGM), including 16 isolates of MAC, were also tested. Omadacycline was active against all RGM species, with MIC50 ranges of 0.004 to 0.25 and 0.06 to 1 μg/ml for 80% and 100% inhibition, respectively. For M. abscessus subsp. abscessus, MIC50s were 0.06 and 0.12 μg/ml with 80% and 100% inhibition, respectively. There was considerable trailing of the omadacycline endpoint with the RGM. MICs of tigecycline exhibited no trailing and were generally within 1 to 2 dilutions of the 100% inhibition omadacycline MICs. While there was no trailing observed in SGM, omadacycline MICs were higher (MIC range, 8 to >16 μg/ml; n = 41), as previously noted with tigecycline. This study supports further research of omadacycline, including clinical trials, for the treatment of RGM infections, especially M. abscessus.
INTRODUCTION
Nontuberculous mycobacterial (NTM) infections are increasing in the United States and globally (1–4). The Mycobacterium abscessus complex and Mycobacterium avium complex (MAC) are the two most frequently encountered NTM complexes/species among clinical laboratories throughout the United States and are some of the most drug-resistant species (5–8).
Only a few antimicrobials, including macrolides (for isolates without a functional erythromycin resistance methylase [erm] gene), cefoxitin, imipenem, amikacin, and tigecycline, have been effective for treatment of M. abscessus complex infections (5). Previous studies have shown the glycylcycline tigecycline to be highly active against rapidly growing mycobacteria (RGM), including tetracycline-resistant isolates (9). Tigecycline has also been shown to be active clinically in an open-label multidrug study (10). However, gastrointestinal (GI) adverse events are common with tigecycline and its intravenous (i.v.)-only formulation often limits its dosage and frequency of usage. For M. abscessus complex, selection of antimicrobials is focused upon the presence or absence of a functional erm gene (5). Antimicrobial treatment regimens for M. abscessus routinely involve multiple injectable antimicrobials, including potentially toxic agents such as amikacin. Amikacin, imipenem, and cefoxitin are not available orally and may cause serious side effects (11). For other NTM, including MAC, treatment usually involves multiple antimicrobials, often also including i.v. agents, and can also be lengthy and difficult to withstand (12–14). Inhaled amikacin is approved (for MAC) for patients experiencing treatment failure (14).
New effective NTM antimicrobials are desperately needed, especially for patients who have pathogens that are or become macrolide or aminoglycoside resistant or for patients who become intolerant due to side effects of the antimicrobials. In previous studies, omadacycline showed pharmacokinetic advantages of higher and sustained concentrations in plasma, lung epithelial lining fluid, and alveolar cells of omadacycline compared to those of tigecycline (15). Moreover, the therapeutic potential of omadacycline is enhanced by its ability to be administered either i.v. or orally (16). In clinical trials, GI side effects are comparable to those in controls, providing a major advantage over tigecycline (15, 17, 18).
Omadacycline is the first agent from the novel class of aminomethylcyclines (17, 19). Its method of action is as a protein synthesis inhibitor which binds to the 30S ribosomal subunit in the mRNA translation complex and inhibits the binding of aminoacyl-tRNA to the mRNA-ribosome complex (17, 19). Omadacycline has shown potent activity against a wide variety of Gram-positive, Gram-negative, and atypical bacterial pathogens (20). Previous in vitro studies have shown that inhibition of protein synthesis occurs even in strains that express tetracycline efflux pumps and ribosomal protection mechanisms. Small studies previously showed activity against some species of RGM, including Mycobacterium fortuitum, Mycobacterium chelonae, and M. abscessus complex (21, 22).
Thus, we undertook this study of the in vitro susceptibility to omadacycline of clinically significant species of NTM, including the most common drug-resistant groups, MAC and the M. abscessus complex.
RESULTS
Omadacycline MICs for RGM ranged from 0.03 to 1 μg/ml, while omadacycline MICs for the slowly growing mycobacteria (SGM) were 0.06 to >16 μg/ml, with MIC50s of 8 to >16 μg/ml. Trailing was problematic with the majority of RGM isolates tested (Fig. 1 and 2). Therefore, omadacycline MICs were read at 80% (the point at which trailing began with little change in subsequent higher dilutions) and 100% inhibition with all of the RGM. For all RGM (n = 70), the range of omadacycline MIC50s was 0.03 to 1 μg/ml at 100% inhibition; omadacycline MICs were one to three concentrations lower (MIC range = 0.004 to 0.5 μg/ml) when read at 80% inhibition.
FIG 1.
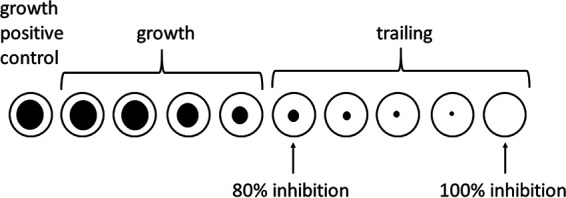
Graphic representation of trailing observed with omadacycline. The wells in which 80% inhibition and 100% inhibition were read are indicated with arrows.
FIG 2.
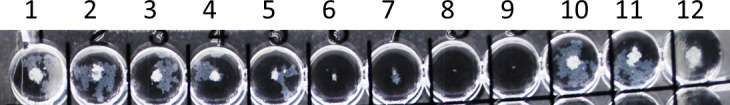
Demonstration of trailing endpoints with omadacycline. Wells 6 and 8 show 80% and 100% inhibition for omadacycline, respectively. Control growth is shown in well 12.
The lowest omadacycline MICs (MIC50 = 0.12 at 100% and 0.015 μg/ml at 80% omadacycline) were seen with the three isolates of M. abscessus subsp. massiliense. Similar omadacycline MIC50 and MIC90s (at 100% inhibition) were noted with M. abscessus subsp. abscessus and the M. fortuitum group. Among the RGM, the highest omadacycline MIC90s were seen with the isolates of M. chelonae, Mycobacterium immunogenum, and the Mycobacterium mucogenicum group (MIC90 = 0.5) (Table 1).
TABLE 1.
MICs of omadacycline and comparator antimicrobials against 70 rapidly growing mycobacteriaa
| Organism (n) | MIC type | MIC (μg/ml) of: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OMC (100% inhibition) |
OMC (80% inhibition) |
DOX | MIN | TGC | AMK | FOX | SXT | LZD | CIP | MXF | IPM | CLR | ||
| M. abscessus subsp. abscessus (20) | Range | 0.06–0.5 | 0.015–0.12 | >8 | 4–>8 | ≤0.015–1 | ≤2–>64 | 32–64 | >4/76 | ≤2–32 | ≥4 | 4–16 | 4–>32 | ≤2–>16 |
| 50% | 0.12 | 0.06 | >8 | >8 | 0.12 | 8 | 32 | >4/76 | 16 | ≥4 | 8 | 8 | ≥16 | |
| 90% | 0.25 | 0.12 | >8 | >8 | 0.25 | 16 | 32 | >4/76 | 32 | ≥4 | 16 | 16 | ≥16 | |
| M. abscessus subsp. massiliense (3) | Range | 0.06–0.25 | 0.015 | >8 | 4–>8 | 0.06–0.25 | 8–32 | 32 | >4/76 | ≤2–16 | ≥4 | 2–16 | 8–32 | ≤2 |
| 50% | 0.12 | 0.015 | >8 | >8 | 0.25 | 8 | 32 | 4/76 | 4 | ≥4 | 4 | 16 | ≤2 | |
| M. chelonae (15) | Range | 0.03–0.5 | 0.004–0.25 | >8 | 4–>8 | 0.015–0.5 | ≤2–32 | >64 | 2/38–>4/76 | ≤2–16 | 0.25–>4 | 1–>16 | 8–>32 | ≤2 |
| 50% | 0.12 | 0.12 | >8 | >8 | 0.25 | 16 | >64 | ≥4/76 | 8 | 2 | 2 | ≥32 | ≤2 | |
| 90% | 0.5 | 0.25 | >8 | >8 | 0.5 | 32 | >64 | ≥4/76 | 16 | ≥4 | 8 | ≥32 | ≤2 | |
| M. goodii (2) | Range | 0.06 | 0.03 | ≤0.12 | ≤0.5 | ≤0.015–0.03 | ≤2 | 32–>64 | ≤1/19 | ≤2 | ≤0.12 | ≤0.06 | ≤2–4 | 16–32 |
| M. wolinskyi (1) | Range | 0.5 | 0.12 | 1 | ≤0.5 | 0.03 | ≤2 | 64 | 2/38 | 4 | 1 | 0.25 | 4 | >32 |
| M. immunogenum (7) | Range | 0.03–0.5 | 0.008–0.5 | 1–>8 | ≤0.5–>8 | 0.03–0.5 | 4–16 | >64 | 2/38–>4/76 | 4–32 | 0.5–>4 | 2–16 | 16–32 | ≤2 |
| 50% | 0.25 | 0.06 | >8 | >8 | 0.12 | 8 | >64 | ≥4/76 | 8 | 2 | 8 | 16 | ≤2 | |
| 90% | 0.5 | 0.5 | >8 | >8 | 0.5 | 16 | >64 | ≥4/76 | 32 | ≥4 | 16 | 32 | ≤2 | |
| M. fortuitum group (12) | Range | 0.06–0.25 | 0.015–0.25 | ≤0.12–>8 | ≤0.5–>8 | ≤0.015–0.12 | ≤2 | ≤16–>64 | ≤1/19–2/38 | ≤2–4 | ≤0.12–0.5 | ≤0.06–0.25 | ≤2–8 | ≤2–>8 |
| 50% | 0.12 | 0.06 | 1 | ≤0.5 | 0.03 | ≤2 | 32 | ≤1/19 | ≤2 | 0.25 | 0.12 | ≤2 | ≥8 | |
| 90% | 0.25 | 0.12 | ≥8 | ≥8 | 0.12 | ≤2 | >64 | 2/38 | 4 | 0.5 | 0.25 | 8 | ≥8 | |
| Doxycycline resistantb (5) | Range | 0.06–0.25 | 0.03–0.25 | 4–>8 | 1–>8 | ≤0.015–0.03 | ||||||||
| 50% | 0.12 | 0.12 | >8 | 4 | 0.03 | |||||||||
| 90% | 0.25 | 0.25 | >8 | >8 | 0.03 | |||||||||
| Doxycycline susceptible (7) | Range | 0.12–0.25 | 0.015–0.12 | ≤0.12–1 | ≤0.5 | ≤0.015–0.12 | ||||||||
| 50% | 0.12 | 0.06 | ≤0.12 | ≤0.5 | 0.03 | |||||||||
| 90% | 0.25 | 0.12 | 1 | ≤0.5 | 0.12 | |||||||||
| M. mucogenicum group (10) | Range | 0.12–1 | 0.03–0.5 | ≤0.12–>8 | ≤0.5–>8 | ≤0.015–0.25 | ≤2 | ≤16 | ≤1/19 | ≤2–4 | ≤0.12–2 | 0.25–1 | ≤2 | ≤2 |
| 50% | 0.25 | 0.12 | 0.25 | ≤0.5 | 0.06 | ≤2 | ≤16 | ≤1/19 | ≤2 | 0.5 | 0.25 | ≤2 | ≤2 | |
| 90% | 0.5 | 0.25 | ≥8 | ≥8 | 0.25 | ≤2 | ≤16 | ≤1/19 | ≤2 | 2 | 0.5 | ≤2 | ≤2 | |
| Doxycycline resistantb (4) | Range | 0.12–0.5 | 0.03–0.25 | 4–>8 | 4–>8 | ≤0.015–0.25 | ||||||||
| 50% | 0.12 | 0.03 | >8 | 4 | 0.06 | |||||||||
| Doxycycline susceptible (6) | Range | 0.25–1 | 0.06–0.5 | ≤0.12–0.25 | ≤0.5 | 0.06–0.25 | ||||||||
| 50% | 0.25 | 0.12 | ≤0.12 | ≤0.5 | 0.06 | |||||||||
| 90% | 1 | 0.5 | 0.25 | ≤0.5 | 0.25 | |||||||||
Because of the small number of isolates tested, M. goodii and M. wolinskyi MIC50 and MIC90 and M. abscessus subsp. massiliense MIC90 were not calculated. Tobramycin MICs are not reported for M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, M. fortuitum group, M. mucogenicum group, M. goodii, and M. wolinskyi per CLSI (36). The tobramycin MIC range, MIC50, and MIC90 for M. chelonae were ≤2 μg/ml and for M. immunogenum were >8 μg/ml. n, number of isolates; OMC, omadacycline; DOX, doxycycline; MIN, minocycline; TGC, tigecycline; AMK, amikacin, FOX, cefoxitin; SXT, trimethoprim-sulfamethoxazole; LZD, linezolid; CIP, ciprofloxacin; MXF, moxifloxacin; IPM, imipenem; CLR, clarithromycin.
Doxycycline-resistant isolates include isolates with intermediate and resistant MICs (≥2 μg/ml).
Among the 12 isolates of the M. fortuitum group, there were five doxycycline-resistant and seven doxycycline-susceptible isolates. The omadacycline MIC50s and MIC90s (0.12 and 0.25 μg/ml, respectively) were the same as for the total 12 isolates in the group (Table 1).
Likewise, four of the 10 isolates of the M. mucogenicum group were doxycycline resistant and six were doxycycline susceptible. The omadacycline MIC50s and MIC90s did not vary by more than one dilution between the two groups (Table 1). Importantly, this finding is much like those of earlier studies with tigecycline (9), which appears to have a similar or the same mechanism of action. Thus, this study shows that omadacycline was also effective in vitro against both doxycycline-resistant and -susceptible isolates of RGM.
Among the SGM tested (n = 41), omadacycline MICs for all species, including MAC, were generally similar, with MIC50s and MIC90s of >16 μg/ml except for five isolates of Mycobacterium kansasii which had an MIC50 of 8 μg/ml and one of the two isolates of Mycobacterium marinum (MIC = 4 μg/ml) (Table 2). Trailing was not a problem with the SGM. Therefore, all omadacycline MIC readings were at 100% inhibition.
TABLE 2.
MICs of omadacycline and comparator antimicrobials against 25 slowly growing mycobacteria other than Mycobacterium avium complexa
| Mycobacterium species (n)b | MIC type | MIC (μg/ml) of: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OMC | DOX | MIN | AMK | SXT | LZD | CIP | MXF | CLR | RFB | RIF | ||
| M. arupense (6) | Range | 1–>16 | 0.5–16 | 0.5–8 | ≤2–64 | ≤0.5/9.5–4/76 | ≤1–32 | >4 | 8–>16 | ≤0.06–4 | ≤0.25–0.5 | 0.5–>2 |
| 50% | >16 | 8 | 8 | 16 | 2/38 | 16 | >4 | >16 | 1 | ≤0.25 | 2 | |
| 90% | >16 | 16 | 8 | 64 | 4/76 | 32 | >4 | >16 | 4 | 0.5 | 2 | |
| M. simiae (7) | Range | >16 | ≥16 | ≥8 | 8–64 | 2/38–4/76 | 16–>32 | 2–>4 | 2–>8 | 2–4 | >2 | >2 |
| 50% | >16 | >16 | ≥8 | 16 | 4/76 | ≥32 | >4 | 4 | 4 | >2 | >2 | |
| 90% | >16 | >16 | ≥8 | 64 | 4/76 | ≥32 | >4 | >8 | 4 | >2 | >2 | |
| M. kansasii (5) | Range | 4–>16 | 0.12–16 | ≤0.25–>8 | ≤2–8 | ≤0.5/9.5–2/38 | ≤1–32 | ≤0.12–4 | ≤0.06–2 | 0.12–1 | ≤0.25 | ≤0.25–>2 |
| 50% | 8 | 4 | 2 | 8 | ≤0.5/9.5 | 2 | 1 | 0.12 | 0.25 | ≤0.25 | 2 | |
| 90% | 16 | 16 | >8 | 8 | 2/38 | 32 | 4 | 2 | 1 | ≤0.25 | >2 | |
| M. marinum (2) | Range | 4–16 | 1–2 | 0.5–1 | ≤2 | ≤0.5/9.5 | ≤1–2 | 4 | 0.5 | 0.25–0.5 | ≤0.25 | 1 |
| M. lentiflavum (3) | Range | 4–>16 | 4–16 | 4–16 | 4 | ≤0.5/9.5–1/19 | 8–16 | 0.5–2 | 0.5–1 | 0.12–0.5 | ≤0.25 | >2 |
| 50% | >16 | 8 | 8 | 4 | 1/19 | 8 | 2 | 1 | 0.25 | >2 | ≤0.25 | |
| M. paraffinicum (2) | Range | 8–>16 | 4–8 | 4 | ≤2–4 | 1/19 | 16–32 | 0.25–>4 | 0.25–1 | 0.25–1 | ≤0.25 | 2 |
OMC, omadacycline; DOX, doxycycline; MIN, minocycline; AMK, amikacin; SXT, trimethoprim sulfamethoxazole; LZD, linezolid; CIP, ciprofloxacin; MXF, moxifloxacin; CLR, clarithromycin; RFB, rifabutin; RIF, rifampin.
The MIC50 and/or MIC90 was not calculated for species with <5 isolates tested.
All other MICs for the comparator antimicrobials were within expected ranges for the taxa of RGM and SGM tested (Tables 2 and 3). Tigecycline MICs were ≤1 μg/ml for all RGM isolates tested. Similar to a previous study (9), tigecycline MIC50s ranged from 0.03 to 0.25 μg/ml, including in the doxycycline-resistant species.
TABLE 3.
MICs of omadacycline and comparator antimicrobials against 16 isolates of Mycobacterium avium complexa
| MIC type | MIC (μg/ml) of: |
||||
|---|---|---|---|---|---|
| OMC | AMK | LZD | MXF | CLR | |
| Range | 0.06–>16 | 4–>64 | 2–>32 | 0.12–4 | ≤0.06–4 |
| 50% | >16 | 8 | 16 | 1 | 1 |
| 90% | >16 | 32 | >32 | 4 | 2 |
OMC, omadacycline; AMK, amikacin; LZD, linezolid; MXF, moxifloxacin; CLR, clarithromycin.
Quality control was performed at each testing event. The CLSI acceptable range of MICs for Staphylococcus aureus ATCC 29213 was 0.12 to 1 μg/ml, and the MIC of omadacycline for Escherichia coli ATCC 25922 was 0.25 μg/ml. All 10 replicates of S. aureus ATCC 29213 and 15 isolates of E. coli ATCC 25922 had an omadacycline MIC within the CLSI acceptable ranges (23). All QC isolates tested with the comparator agents was within the CLSI acceptable range for Mycobacterium peregrinum ATCC 700686, M. marinum ATCC 927, and S. aureus ATCC 29213. Ten replicates of M. peregrinum ATCC 700686 had an MIC range for omadacycline of 0.06 to 0.12 μg/ml (mode = 0.12 μg/ml). Nine replicates of M. abscessus ATCC 19977T were tested and had an omadacycline MIC range of 0.25 to 0.5 μg/ml (mode = 0.5 μg/ml). Eight replicates of M. marinum ATCC 927 had an omadacycline MIC range of 2 to 4 μg/ml. One isolate of M. avium ATCC 700898 had an omadacycline MIC of 8 μg/ml.
DISCUSSION
Previous small studies showed in vitro activity of omadacycline against RGM, including M. chelonae, the M. fortuitum group, and the difficult-to-treat isolates of the M. abscessus complex (21, 22, 24). To our knowledge, no in vitro MIC studies with omadacycline against other species of NTM have been published.
Previous MIC studies reported omadacycline MIC50s of 1 and 2 μg/ml, respectively, for isolates of the M. abscessus complex (combining M. abscessus subsp. abscessus and M. abscessus subsp. massiliense) with species identification based upon erm gene sequence. In the study by Kaushik and colleagues, the MIC50 and MIC90s for M. abscessus subsp. abscessus were one dilution higher than those for the combined grouping of M. abscessus subsp. abscessus and M. abscessus subsp. massiliense (Table 4) (21, 22). The M. abscessus complex MICs determined by Shoen and colleagues were read at 5 days after incubation at 37°C. As stated previously, the MICs in the present study, except for a single isolate of M. chelonae, were read at 3 days incubation at 30°C as per CLSI recommendations (25). It is known that the tetracyclines in general are unstable after extended incubation, and thus, these previously reported MICs may be higher than our MICs due to their longer incubation (11). Shoen et al. also tested 22 isolates of M. chelonae at 4 days after incubation at 32°C. These omadacycline MIC50 and MIC90s were 0.125 and 0.25 μg/ml, respectively, and similar to our MICs. These MICs were similar to those for our isolates of the M. fortuitum group (MIC50 and MIC90s of 0.125 and 0.5 μg/ml compared to 0.12 and 0.25 μg/ml, respectively) which they also tested. However, MICs for these isolates were read after only 48 h incubation in their study (21).
TABLE 4.
Comparison of three studies of rapidly growing mycobacteria against omadacycline and tigecycline
| Study | No. of isolates tested | Incubation temp/time | Species or complex | MIC (μg/ml) of: |
|||
|---|---|---|---|---|---|---|---|
| Omadacycline |
Tigecycline |
||||||
| 50% | 90% | 50% | 90% | ||||
| Shoen et al. (21) | 24 | 37°C/5 days | M. abscessus subsp. abscessus | 1 | 2 | 1 | 2 |
| 22 | 32°C/4 days | M. chelonae | 0.125 | 0.25 | 0.06 | 0.25 | |
| 20 | 37°C/2 days | M. fortuitum group | 0.125 | 0.5 | 0.25 | 0.5 | |
| Kaushik et al. (22) | 16 | 30°C/3 days | M. abscessus subsp. abscessus | 2 | 4 | 1 | 2 |
| 12 | 30°C/3 days | M. abscessus subsp. massiliense and M. abscessus subsp. massiliense-bolletii | 1 | 2 | 1 | 2 | |
| This study | 20 | 30°C/3 days | M. abscessus subsp. abscessus | 0.12 (0.06) | 0.25 (0.12) | 0.12 | 0.25 |
| 3 | 30°C/3 days | M. abscessus subsp. massiliense | 0.12 (0.015) | 0.25 | |||
| 15 | 30°C/3 days | M. chelonae | 0.12 (0.12) | 0.5 (0.25) | 0.25 | 0.5 | |
| 12 | 30°C/3 days | M. fortuitum group | 0.12 (0.06) | 0.25 (0.12) | 0.03 | 0.12 | |
A separate report of testing the type strain of M. abscessus subsp. abscessus (CIP 104536, ATCC 19977) in the Netherlands using CLSI guidelines showed omadacycline and tigecycline MICs of 4 μg/ml when the strain was tested in duplicate at 35°C in cation-adjusted Mueller-Hinton broth (24). These MICs are in contrast to an MIC of 1 μg/ml for both agents reported by Kaushik and colleagues and our finding of omadacycline MICs of 0.25 to 0.5 μg/ml with this strain. The tigecycline MIC range for ATCC 19977T in our laboratory was 0.12 to 0.5 μg/ml.
The two previous studies showed tigecycline and omadacycline MIC50 and MIC90s to be 3 to 4 dilutions higher than those in the present study. The differences between this study and the study by Shoen et al. may be explained, as noted above, by the longer incubation and differences in incubation temperatures in their study (21). However, the differences between the study by Kaushik et al. and this study cannot be attributed to differences in incubation parameters, since their incubation times and temperatures were the same as ours. Instead, possible reasons for differences could be related to the study population or geographical origin of the isolates (22). Furthermore, there was no mention of trailing endpoints in either study, and these higher MICs could have been related to that problem, as we noted with the omadacycline MICs in the present study.
The current CLSI document (25) states that mycobacterial colonies should be transferred to tubes of sterile water containing glass beads. The suspension should be vortexed and large clumps allowed to settle with the supernatant used for the inoculum to avoid clumping of the organisms in broth. Clumping may also cause trailing, leading to higher MIC reads (Fig. 1 and 2).
It should also be noted that our research reference laboratory at the University of Texas Health Science Center at Tyler (UTHSCT) has more than 40 years of experience in antimicrobial susceptibility testing of NTM, with more than 20 years of experience in the susceptibility testing of tigecycline. Moreover, our lower tigecycline MICs have been corroborated by other investigators, including a 2008 study in Spain (26). During this time, we have rarely seen isolates of RGM with tigecycline MICs of >1 μg/ml with our clinical isolates. Our MICs in this study are similar to those previously published by our laboratory and in the Spanish study (9, 26).
Isolates with trailing endpoints present a problem in reading/interpretation of MICs. Since the CLSI has not yet addressed MICs of omadacycline, we recommend that extreme care be taken in reading the MICs, disregarding obvious trailing, which is usually seen as a “ghost-like” or faint button exhibiting much less growth than the positive control. Just as is recommended for difficult-to-read MIC wells, including MICs for trimethoprim-sulfamethoxazole, which are also read at 80% inhibition, we also recommend adjusting the MIC panel on the mirrored light box with overhead illumination so that the wells in which the trailing occurs are in the most brightly lit area.
This in vitro study showed activity of omadacycline against RGM, including isolates of M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, M. chelonae, M. immunogenum, the M. fortuitum group, the M. mucogenicum group, Mycobacterium goodii, and Mycobacterium wolinskyi. Trailing was common among most RGM isolates tested, and thus, we read those MICs at 80% inhibition (ignoring faint buttons of growth, much as trimethoprim-sulfamethoxazole MICs are read), and 100% inhibition. The 80% inhibitory MICs are given in Table 1. Upon careful analysis, we noted that these 80% reads usually varied by ≤2 concentrations. Moreover, the omadacycline MIC50s and MIC90s with both 80% and 100% inhibition were ≤0.5 μg/ml against all of the RGM tested except for six isolates of the M. mucogenicum group, with an MIC90 of 1 μg/ml at 100% inhibition and 0.5 μg/ml at 80% inhibition.
MICs for omadacycline were generally comparable to the tigecycline MICs for most RGM isolates (Table 1). However, unlike with omadacycline, no trailing was obvious with tigecycline. Tigecycline MIC50s were within one dilution of the omadacycline MIC50 (100% inhibition) except for the MIC50s of the M. fortuitum group and the M. mucogenicum group, which were two dilutions less, with tigecycline MIC50s of 0.03 μg/ml and 0.06 μg/ml, respectively, compared to omadacycline MIC50s of 0.12 μg/ml and 0.25 μg/ml, respectively (Table 1). Furthermore, as shown in Table 1, and similar to our earlier findings in the in vitro study of tigecycline, the omadacycline MIC50s and MIC90s were the same or within one dilution for isolates of both doxycycline-resistant and doxycycline-susceptible isolates of the M. fortuitum group and the M. mucogenicum group, respectively.
Although there was no trailing of MICs with the omadacycline with the SGM, omadacycline MICs were similar to our previous tigecycline MIC against the SGM (9). Omadacycline MIC50s and MIC90s were higher than those of the RGM, and MICs were not within the CLSI-recommended MIC range for doxycycline-susceptible isolates (9, 25). The MAC isolates in this study had omadacycline MIC50s of >16 μg/ml (Table 3), and the other SGM exhibited omadacycline MIC50s of ≥8 μg/ml, although fewer than 10 isolates of each species other than MAC were tested (Table 2). Additional experiments to elucidate reasons for the higher MICs with omadacycline with the SGM were beyond the scope of this study.
Previous structural studies of omadacycline demonstrated that the aminomethyl substituent modification at the C-9 position on the tetracycline molecule allows omadacycline to overcome the usual tetracycline resistance mechanisms, including ribosomal protection proteins and efflux mechanisms that characterize the older tetracyclines (16, 17, 19, 27). Tigecycline, a glycylcycline tetracycline, is also known to circumvent these common mechanisms (20). However, since tigecycline is available only for parenteral use and its use is limited by serious adverse events, including nausea, vomiting, and increased mortality related to dosage (10, 20, 28), omadacycline provides an attractive oral option, although it can also be administered intravenously (29). Moreover, the most frequently observed GI adverse events, including nausea and vomiting, were only 14.9 and 8.3%, respectively, in a recent safety study of omadacycline with both oral and intravenous dosing (30).
In a 2017 comparative study of intravenous omadacycline (100 mg every 12 h for two doses followed by 100 mg every 24 h for six doses) and 100 mg tigecycline given i.v. followed by 50 mg every 12 h for six doses in healthy nonsmoking male and female adult subjects, the incidence of nausea with omadacycline was 2.4%, compared to 47.6% in subjects treated with tigecycline. There were no incidences of vomiting with omadacycline, compared to 14.3% in subjects to whom tigecycline was administered. Also, 9.5% of the subjects given tigecycline discontinued treatment because of nausea, compared to 0% of subjects given omadacycline (15).
In the same healthy-subject study, the pattern and time course of tigecycline concentration in plasma, epithelial lining fluid, and alveolar cells were similar to those seen in subjects given omadacycline. However, the tigecycline concentrations were lower than those of omadacycline, suggesting pharmacokinetic advantages of higher and sustained concentrations of omadacycline compared to tigecycline (15). Plasma levels of omadacycline have previously shown a maximum concentration in serum (Cmax) of 2.12 ± 0.68 μg/ml compared to a tigecycline Cmax of 0.98 ± 0.21 μg/ml following a 100-mg dose of omadacycline and a 50-mg dose of tigecycline (15).
Omadacycline was approved by the Food and Drug Administration (FDA) in 2018 for use in acute bacterial skin and skin structure infections and community-acquired bacterial pneumonia (27, 31). Although no indication for mycobacterial infections has been FDA approved, its use against NTM infections is promising. In vitro MIC studies against NTM and the safety and efficacy profile of omadacycline suggest a potential efficacious addition to the RGM treatment armamentarium.
Previous genomic analysis of the M. abscessus complex has revealed the presence of many potential antimicrobial resistance determinants, including multiple tetracycline efflux genes (32). Additional studies with different species of NTM are needed to assess the activity of omadacycline against isolates that express the antibiotic resistance mechanisms that have been previously reported to be overcome by omadacycline, such as resistance genes for ribosomal protection, including tetM, tetO, and tetS, and tetracycline efflux genes, including tetK and tetL (17, 29). Further research with omadacycline focusing on tetracycline resistance mechanisms and more dynamic models, such as animal models and/or the hollow fiber model, may be warranted.
MATERIALS AND METHODS
MIC testing was performed in cation-adjusted Mueller-Hinton broth (CAMHB) as described and recommended by the Clinical and Laboratory Standards Institute (CLSI) (25). Comparator antimicrobials included amikacin (AMK), cefoxitin (FOX), ciprofloxacin (CIP), clarithromycin (CLR), doxycycline (DOX), imipenem (IPM), linezolid (LZD), minocycline (MIN), moxifloxacin (MXF), tigecycline (TGC), and trimethoprim sulfamethoxazole (TMP-SMX) for the RGM (n = 70). Comparator antimicrobials included amikacin, clarithromycin, linezolid, and moxifloxacin for the MAC (n = 16) and amikacin, ciprofloxacin, clarithromycin, doxycycline, linezolid, moxifloxacin, rifabutin, rifampin, and TMP-SMX for the other SGM (n = 25) (Tables 1 to 3) (25).
Species identification.
Isolates of NTM were identified by gene sequencing as indicated for each species/group as previously described. For the RGM, sequencing of region 5 of the rpoβ gene and the erm(41) gene (for the M. abscessus complex) was performed using previously recommended criteria for identification, including CLSI recommendations (33, 34). The SGM species were identified using partial 16S rRNA gene sequencing along with the CLSI interpretive criteria (33, 35).
Antimicrobial susceptibility testing.
Seventy isolates of RGM were tested in CAMHB with incubation at 30°C for 3 days (one isolate of M. chelonae required 6 days for adequate growth in broth). These included six each of doxycycline-susceptible and doxycycline-resistant isolates of the M. fortuitum group and four of 10 doxycycline-resistant isolates of the M. mucogenicum group. Additionally, 41 SGM MICs were determined in CAMHB containing oleic acid-albumin-dextrose-catalase (OADC) and incubated at 35°C for 7 to 8 days. All MIC testing was performed in accordance with the current CLSI guidelines (25, 36).
Quality control.
Quality control (QC) was performed with each susceptibility testing event. QC for RGM and SGM comparator antimicrobials was performed using CLSI guidelines for interpretive criteria for acceptable ranges with M. peregrinum ATCC 700686, Staphylococcus aureus ATCC 29213, and Escherichia coli ATCC 25922 (25, 36).
Omadacycline QC was performed using the same three QC isolates as for comparator agents, including S. aureus ATCC 29213 10 times, and E. coli ATCC 25922 15 times. The omadacycline QC with M. peregrinum ATCC 700686 was performed with 10 replicates. In addition, QC was performed with M. marinum ATCC 927 eight times, M. avium ATCC 700898 one time, and M. abscessus ATCC 19977T nine times.
ACKNOWLEDGMENTS
Funding for this study was provided by Paratek Pharmaceuticals.
A special thank-you goes to Alisa Serio (Paratek Pharmaceuticals), who supported our study and reviewed the manuscript. We thank our coworkers at the UTHSCT, including Adrian Almodovar, Taylor Britten, Indrani Das, Bibiana Gonzalez-Ramirez, Elena Iakhiaeva, and Sruthi Vasireddy for sequencing the isolates, and Georgie Bush, Dolores Hughes, Erica Mabry, Chetna Patel, and Eliana Rodriguez for performing the MIC determinations. We also thank Kavya Somayaji and Mary Davis for their laboratory assistance and Joanne Woodring for her clerical support.
REFERENCES
- 1.Baldwin SL, Larsen SE, Ordway D, Cassell G, Coler RN. 2019. The complexities and challenges of preventing and treating nontuberculous mycobacterial diseases. PLoS Negl Trop Dis 13:e0007083. doi: 10.1371/journal.pntd.0007083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brode SK, Marchand-Austin A, Jamieson FB, Marras TK. 2017. Pulmonary versus nonpulmonary nontuberculous mycobacteria, Ontario, Canada. Emerg Infect Dis 23:1898–1901. doi: 10.3201/eid2311.170959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honda JR, Virdi R, Chan ED. 2018. Global environmental nontuberculous mycobacteria and their contemporaneous man-made and natural niches. Front Microbiol 9:2029. doi: 10.3389/fmicb.2018.02029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivero-Lezcano OM, González-Cortés C, Mirsaeidi M. 2019. The unexplained increase of nontuberculous mycobacteriosis. Int J Mycobacteriol 8:1–6. doi: 10.4103/ijmy.ijmy_18_19. [DOI] [PubMed] [Google Scholar]
- 5.Brown-Elliott BA, Vasireddy S, Vasireddy R, Iakhiaeva E, Howard ST, Nash KA, Parodi N, Strong A, Gee M, Smith T, Wallace RJ, Jr.. 2015. Utility of sequencing the erm(41) gene in isolates of Mycobacterium abscessus subsp. abscessus with low and intermediate clarithromycin MICs. J Clin Microbiol 53:1211–1215. (Erratum, 54:1172, 2016.) doi: 10.1128/JCM.02950-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Jr, Winthrop K, Infectious Disease Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 7.Koh WJ, Stout JE, Yew WW. 2014. Advances in the management of pulmonary disease due to Mycobacterium abscessus complex. Int J Tuber Lung Dis 18:1141–1148. doi: 10.5588/ijtld.14.0134. [DOI] [PubMed] [Google Scholar]
- 8.Brown-Elliott BA, Iakhiaeva E, Griffith DE, Woods GL, Stout JE, Wolfe CR, Turenne CY, Wallace RJ, Jr. 2013. In vitro activity of amikacin against isolates of Mycobacterium avium complex with proposed MIC breakpoints and finding of a 16S rRNA gene mutation in treated isolates. J Clin Microbiol 51:3389–3394. (Erratum, 52:1311, 2014.) doi: 10.1128/JCM.01612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace RJ, Jr, Brown-Elliott BA, Crist CJ, Mann L, Wilson RW. 2002. Comparison of the in vitro activity of the glycylcycline tigecycline (formerly GAR-936) with those of tetracycline, minocycline, and doxycycline against isolates of nontuberculous mycobacteria. Antimicrob Agents Chemother 46:3164–3167. doi: 10.1128/AAC.46.10.3164-3167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace RJ, Jr, Dukart G, Brown-Elliott BA, Griffith DE, Scerpella EG, Marshall B. 2014. Clinical experience in 52 patients with tigecycline-containing regimens for salvage treatment of Mycobacterium abscessus and Mycobacterium chelonae infections. J Antimicrob Chemother 69:1945–1953. doi: 10.1093/jac/dku062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown-Elliott BA, Nash KA, Wallace RJ, Jr.. 2012. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin Microbiol Rev 25:545–582. doi: 10.1128/CMR.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koh WJ, Jeon K, Lee NY, Kim B-J, Kook Y-H, Lee S-H, Park Y-K, Kim CK, Shin SJ, Huitt GA, Daley CL, Kwon OJ. 2011. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am J Respir Crit Care Med 183:405–410. doi: 10.1164/rccm.201003-0395OC. [DOI] [PubMed] [Google Scholar]
- 13.van Ingen J, Boeree MJ, van Soolingen D, Mouton JW. 2012. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist Updat 15:149–161. doi: 10.1016/j.drup.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Griffith DE, Eagle G, Thomson R, Aksamit TR, Hasegawa N, Morimoto K, Addrizzo-Harris DJ, O’Donnell AE, Marras TK, Flume PA, Loebinger MR, Morgan L, Codecasa LR, Hill AT, Ruoss SJ, Yim J-J, Ringshausen FC, Field SK, Philley JV, Wallace RJ, van Ingen J, Coulter C, Nezamis J, Winthrop KL. 2018. Amikacin liposome inhalation suspension for treatment-refractory lung disease caused by Mycobacterium avium complex (CONVERT). A prospective, open-label, randomized study. Am J Respir Crit Care Med 198:1559–1569. doi: 10.1164/rccm.201807-1318OC. [DOI] [PubMed] [Google Scholar]
- 15.Gotfried MH, Horn K, Garrity-Ryan L, Villano S, Tzanis E, Chitra S, Manley A, Tanaka SK, Rodvold KA. 2017. Comparison of omadacycline and tigecycline pharmacokinetics in the plasma, epithelial lining fluid, and alveolar cells of healthy adult subjects. Antimicrob Agents Chemother 61:e01135-17. doi: 10.1128/AAC.01135-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opal S, File TM, Jr, van der Poll T, Tzanis E, Chitra S, McGovern PC. 2019. An integrated safety summary of omadacycline, a novel aminomethylcyclines antibiotic. Clin Infect Dis 69:S40–S47. doi: 10.1093/cid/ciz398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka SK, Steenbergen J, Villano S. 2016. Discovery, pharmacology, and clinical profile of omadacycline, a novel aminomethylcyclines antibiotic. Bioorg Med Chem 24:6409–6419. doi: 10.1016/j.bmc.2016.07.029. [DOI] [PubMed] [Google Scholar]
- 18.Bundrant LA, Tzanis E, Garrity-Ryan L, Bai S, Chitra S, Manley A, Villano S. 2017. Safety and pharmacokinetics of the aminomethylcyclines antibiotic omadacycline administered to healthy subjects in oral multiple-dose regimens. Antimicrob Agents Chemother 62:e1487-17. doi: 10.1128/AAC.01487-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidrich C, Mitova S, Schedlbauer A, Connell S, Fucini P, Steenbergen J, Berens C. 2016. The novel aminomethylcycline omadacycline has high specificity for the primary tetracycline-binding site on the bacterial ribosome. Antibiotics (Basel): 5:e32. doi: 10.3390/antibiotics5040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villano S, Steenbergen J, Loh E. 2016. Omadacycline: development of a novel aminomethylcycline antibiotic for treating drug-resistant bacterial infections. Future Microbiol 11:1421–1434. doi: 10.2217/fmb-2016-0100. [DOI] [PubMed] [Google Scholar]
- 21.Shoen C, Benaroch D, Sklaney M, Cynamon M. 2019. In vitro activities of omadacycline against rapidly growing mycobacteria. Antimicrob Agents Chemother 63:e02522-18. doi: 10.1128/AAC.02522-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaushik A, Ammerman NC, Martins O, Parrish NM, Nuermberger EL. 2019. In vitro activity of new tetracycline analogs omadacycline and eravacycline against drug-resistant clinical isolates of Mycobacterium abscessus. Antimicrob Agents Chemother 63:e00470-19. doi: 10.1128/AAC.00470-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing, 30th ed. CLSI document M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 24.Bax HI, de Vogel CP, Mouton JW, de Steenwinkel JEM. 2019. Omadacycline as a promising new agent for the treatment of infections with Mycobacterium abscessus. J Antimicrob Chemother 74:2930–2933. doi: 10.1093/jac/dkz267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2018. Susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes, 3rd ed. CLSI standard document M24. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 26.Fernández-Roblas R, Martín-de-Hijas NZ, Fernández-Martínez AI, García-Almeida D, Gadea I, Esteban J. 2008. In vitro activities of tigecycline and 10 other antimicrobials against nonpigmented rapidly growing mycobacteria. Antimicrob Agents Chemother 52:4184–4186. doi: 10.1128/AAC.00695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stets R, Popescu M, Gonong JR, Mitha I, Nseir W, Madej A, Kirsch C, Das AF, Garrity-Ryan L, Steenbergen JN, Manley A, Eckburg PB, Tzanis E, McGovern PC, Loh E. 2019. Omadacycline for community-acquired bacterial pneumonia. N Engl J Med 380:517–527. doi: 10.1056/NEJMoa1800201. [DOI] [PubMed] [Google Scholar]
- 28.Tasina E, Haidich AB, Kokkali S, Arvanitidou M. 2011. Efficacy and safety of tigecycline for the treatment of infectious diseases: a meta-analysis. Lancet Infect Dis 11:834–844. doi: 10.1016/S1473-3099(11)70177-3. [DOI] [PubMed] [Google Scholar]
- 29.Karlowsky JA, Steenbergen J, Zhanel GG. 2019. Microbiology and preclinical review of omadacycline. Clin Infect Dis 69:S6–S15. doi: 10.1093/cid/ciz395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher JC. 2019. Omadacycline: a modernized tetracycline. Clin Infect Dis 69:S1–S5. doi: 10.1093/cid/ciz394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chambers HF. 2019. Omadacycline—the newest tetracycline. N Engl J Med 380:588–589. doi: 10.1056/NEJMe1900188. [DOI] [PubMed] [Google Scholar]
- 32.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrman J-L, Daffe M, Brosch R, Risler J-L, Gaillard JL. 2009. Non-mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2018. Interpretive criteria for identification of bacteria and fungi by targeted DNA sequencing, 3rd ed. CLSI guideline MM18. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 34.Adékambi T, Colson P, Drancourt M. 2003. rpo B-based identification of nonpigmented and late pigmented rapidly growing mycobacteria. J Clin Microbiol 41:5699–5708. doi: 10.1128/JCM.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards U, Rogall T, Blöcker H, E M, Böttger EC. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. 2018. Performance standards for susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes. CLSI document M62. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]


