FIG 10.
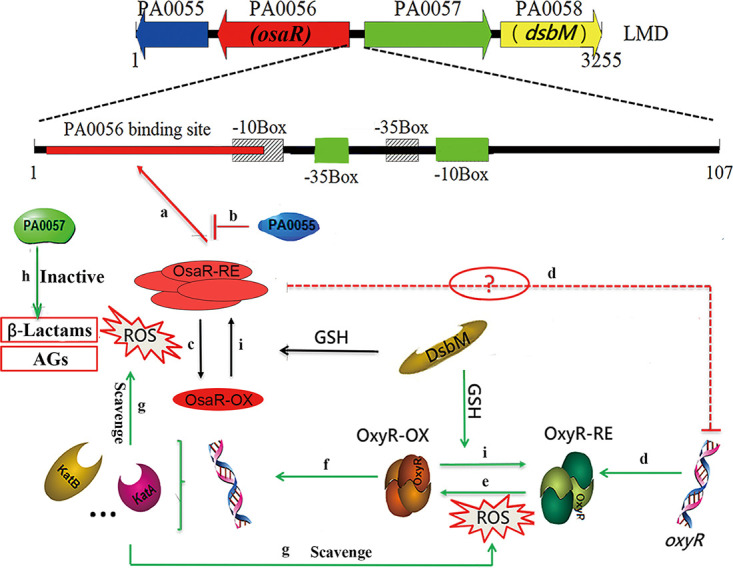
Proposed model for oxidative stress responses regulated by OsaR in P. aeruginosa. In this hypothetical model, the bacteria have a reduced internal milieu under normal physiological conditions. Therefore, reduced OsaR protein binds to its binding site in the form of a polymer, which facilitates transcriptional initiation of osaR (a). The negative feedback regulation of PA0055 protein keeps the positive autoregulation of osaR at a relative level (b). The transcription of PA0057 and dsbM is normally repressed. When exposed to antibiotics (β-lactams or AGs) or oxidative stress, OsaR is oxidized and forms monomers (c) and OxyR is activated (e), resulting in increased expression of antioxidant genes to restore the redox balance. OsaR protein is oxidized, forming a disulfide bond, and is converted into a nonbinding state (c). The expression levels of PA0055 and osaR are downregulated, but PA0057, dsbM, and oxyR (d) (the dotted line indicates indirect regulation) are upregulated. Among them, the metallo-β-lactamase expressed by PA0057 protein can hydrolyze the lactamide ring of β-lactams and enhance the tolerance of P. aeruginosa to lactams (h). Oxidized OxyR can continuously activate the transcription of downstream antioxidant genes to scavenge ROS (f and g). Thus, the influence of AGs on bacteria can be alleviated. Oxidized OxyR and oxidized OsaR could be catalyzed to a reduced state by DsbM when the oxidation stress is released (i). Finally, the LMD gene cluster is restored to the initial physiological state. The model marked with green lines integrates previous published data.
