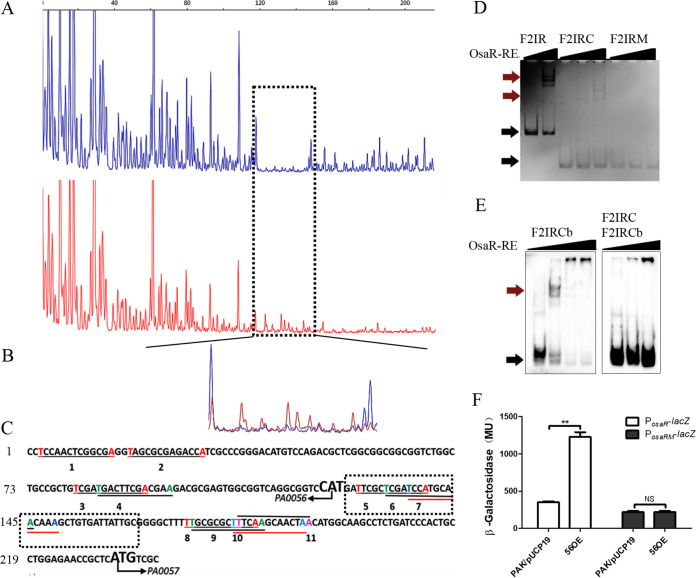
FIG 8.
Identification of the OsaR-protected cis elements in the IR promoter region using a DNase I footprinting assay and EMSA. (A and B) The probes used were labeled with FAM dye, and the control products (red) together with the digestion products (blue) were analyzed with peak scanner software v1.0 (Applied Biosystems) (A). These two sequencing results are indicated by different colors separately and are then merged together (B). The whole region of the coding strand DNA is shown in panel A, and the region protected by OsaR from DNase I cleavage was enlarged and is shown separately in panel B. (C) The region protected by OsaR from DNase I digestion is the OsaR binding site, which is in the black dotted box; other salient features named 1 to 13 are all T-N11-A boxes in the DNA fragment identified by a program. (D) F2IR was cut from 238 bp to 90 bp (designated F2IRC); in the F2IRC site mutant, the four bases in bold are mutation sites (designated F2IRM). EMSAs of OsaR-RE binding with the site mutant DNA fragment F2IRM were performed; F2IR and F2IRC are positive controls, and OsaR-RE concentrations were 0 nM, 200 nM, or 400 nM. (E) For cold competition, OsaR-RE was first incubated with the DNA fragment F2IRC and then F2IRC marked with biotin (F2IRCb) was added. The OsaR-RE concentrations were 0 nM, 200 nM, and 400 nM, F2IRCb was a positive control, with OsaR-RE concentrations of 0 nM, 200 nM, 400 nM, and 800 nM. (F) Promoter activity was analyzed in the WT strain and OsaR overexpression strains, with four base mutations of the osaR promoter PosaRM in the putative OsaR binding site as F2IRM. The PosaR-lacZ reporter was a control. Red arrows indicate the protein-DNA complex, and black arrows indicate free DNA. NS, not significant. **, P < 0.01 by Student's t test.