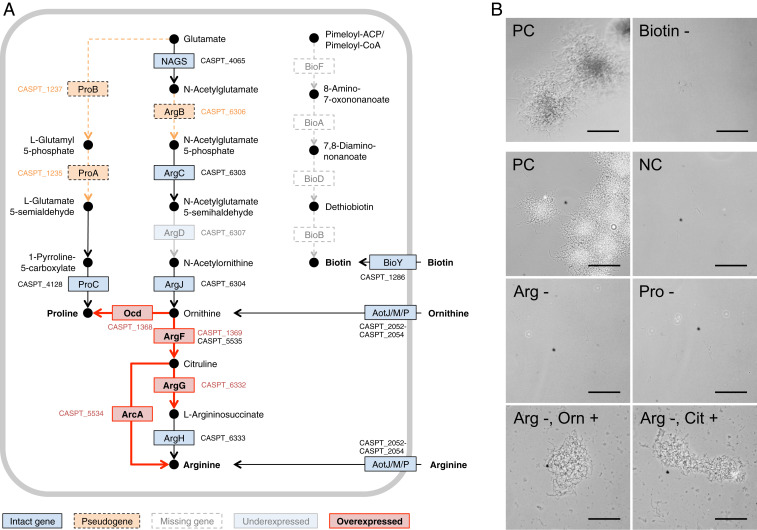
Fig. 4.
Effect of genome erosion on amino acid requirements of “S. philanthi biovar triangulum” strain 23Af2. (A) Proline, arginine, and biotin biosynthetic pathways. Pseudogenes are indicated in an orange color, and dashed lines denote putatively interrupted enzymatic steps. Missing genes are indicated in white boxes. Intact genes with no differential expression between antenna and in vitro cultures are in blue. Genes overexpressed (log2 > 1) in antennal samples are highlighted in red, and those underexpressed (log2 < −1) are in light blue. All differentially expressed genes shown had an adjusted P value (q-value) < 0.1, with the exception of CSP_1369. (B) Phase-contrast microscopy images of in vitro cultures of “S. philanthi biovar triangulum” without biotin, arginine, or proline show no growth. Arginine auxotrophy can be bypassed by the addition of precursors ornithine (Orn) or citrulline (Cit). NC, negative control (Grace’s based medium without added amino acids or vitamins); PC, positive control (complete Grace’s medium); Arg, arginine; Pro, proline. (Scale bar corresponds to 100 µm.)