FIG 6.
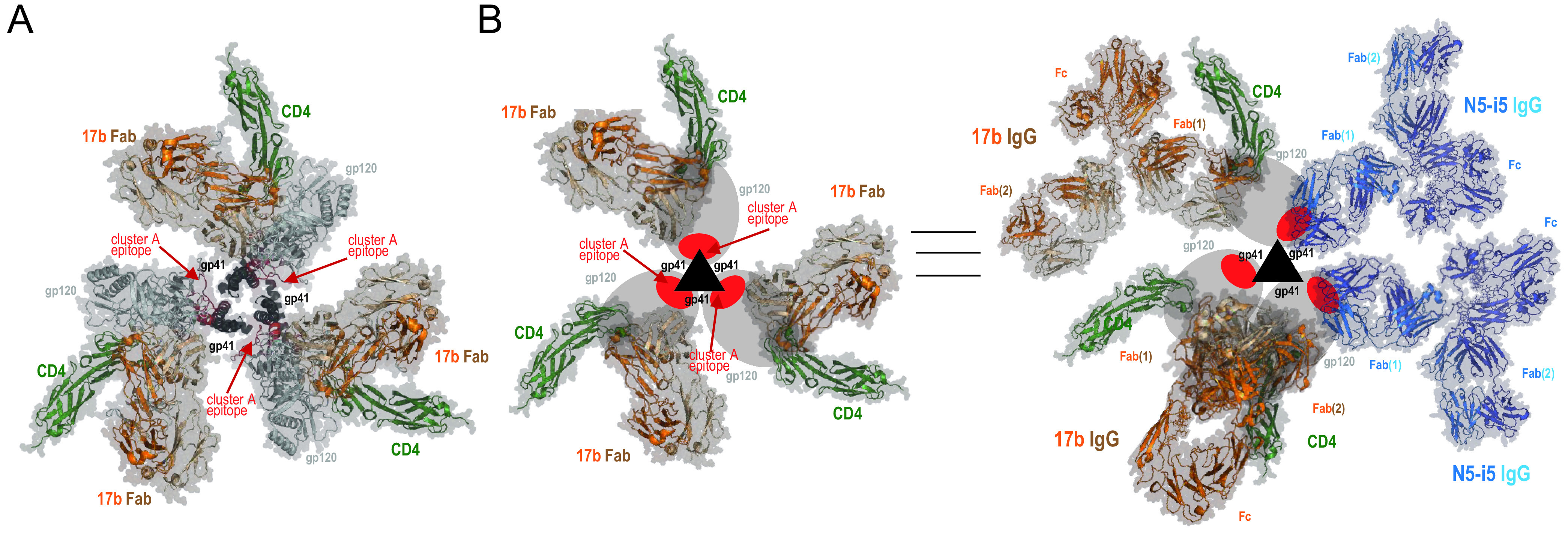
Model of asymmetric HIV-1 trimer opening induced by binding of soluble CD4 and CoRBS-specific antibody 17b. (A) Env trimer–CD4-17b Fab complex as in the structure under PDB accession number 3J70 (30). The epitope footprint of anti-cluster A antibody N5-i5 is highlighted in red. (B) Model of the asymmetric opening of the Env trimer upon binding to soluble CD4 and IgG of CoRBS-specific antibody 17b. Env trimer promoters are shown schematically, gp120 is shown as ovals, and gp41 is shown as triangles. The cluster A epitope region is highlighted in red. (Left) Env trimer–CD4-17b Fab complex as described above for panel A. The cluster A region is buried at the trimer interface and not accessible for antibody recognition. (Right) Structural rearrangements of the Env trimer upon CD4 and 17b IgG binding. The model involves two 17b IgG molecules bound to the CD4-triggered HIV-1 trimer, one with both Fab arms involved in an intraprotomer interaction and a second IgG interacting through one Fab arm only. The cluster A region becomes exposed by rearrangements of the gp120 subunits relative to gp41 in the trimer to expose two binding sites for N5-i5 IgG. Based on the binding data, two N5-i5 IgGs are bound per trimer (attached by a single Fab arm).
