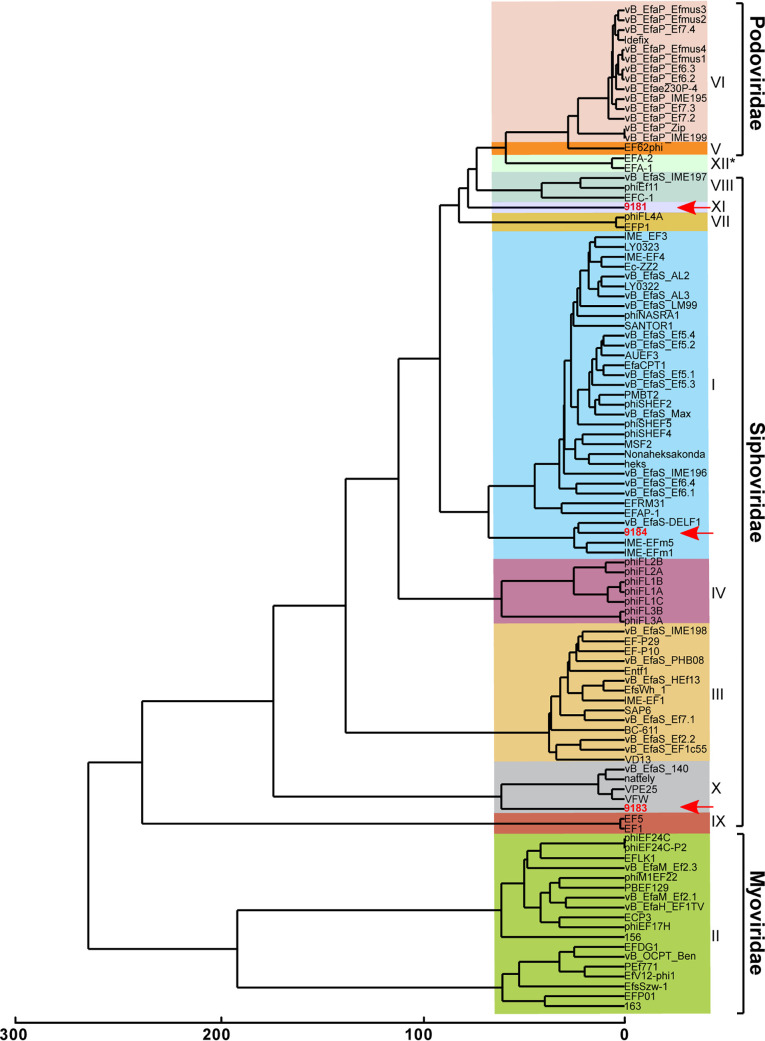
FIG 2.
Comparative genomic analysis identifies two novel enterococcal phage orthoclusters. A comparative genome analysis was performed using OrthoMCL as described previously by Bolocan et al. (16). A phylogenetic proteomic tree was constructed from the OrthoMCL matrix using the Manhattan distance metric and hierarchical clustering using an average linkage with 1,000 iterations. Ninety-nine enterococcal phage genomes available from NCBI were used for comparison to E. faecium phages 9181, 9183, and 9184 (highlighted in red, emphasized by red arrows). Distinct phage orthoclusters are represented by colored boxes. Roman numerals to the right of the shaded boxes signify the phage orthocluster number. Phage orthocluster morphology is indicated by calipers (if known) or an asterisk symbol (if unknown) to the right of the roman numerals.