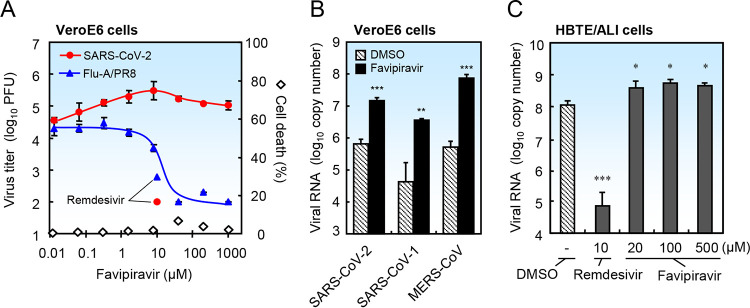
FIG 1.
Favipiravir does not block replication of SARS-CoV-2 in cultured cells. To minimize the effects of the drug solvent, 400 mM favipiravir (23384, Cayman Chemical) was prepared in dimethyl sulfoxide (DMSO) as a stock solution and diluted >400-fold in medium before use. (A) VeroE6 cells seeded in 96-well plates were infected with SARS-CoV-2 (strain WK-521) or influenza A virus (strain PR8) at an MOI (multiplicity of infection) of 0.1 in the presence of DMSO or favipiravir. To prime influenza virus and SARS-CoV-2 for infection, 1 μg/ml trypsin was added to the medium. After incubation for 2 days, the culture media were collected and the virus titer of SARS-CoV-2 or influenza virus was measured by a plaque assay using VeroE6/TMPRSS2 cells (5) or MDCK cells, respectively. Data represent the average of three independent experiments (n = 3). Average cell death in the absence of virus was measured in a WST assay (n = 4). (B) VeroE6 cells were infected with SARS-CoV-2 (strain WK-521), SARS-CoV-1 (strain Frankfurt), or MERS-CoV (strain EMC) at an MOI of 0.1 in the presence of DMSO or favipiravir (8 μM), and then incubated for 2 days. Trypsin was not added to the culture medium. Viral RNA was extracted from the culture medium and quantified by real-time PCR using the SARS-2-E, SARS-N, and MERS-upE primer/probe sets (n = 4) (11, 12). (C) Differentiated human bronchial tracheal epithelial cells (HBTE/ALI cells) were infected with SARS-CoV-2 at an MOI of 0.01 in the presence of DMSO or favipiravir, and then incubated for 3 days. Viral RNA was extracted from cells and quantified by real-time PCR using the SARS-2-E primer/probe set (11). Data are presented as the mean ± standard deviation (SD) (n = 4). Two-tailed Student’s t tests were used to analyze statistical significance compared with the DMSO control: *, significant (P ≤ 0.05); **, highly significant (P ≤ 0.01); and ***, very highly significant (P ≤ 0.001).